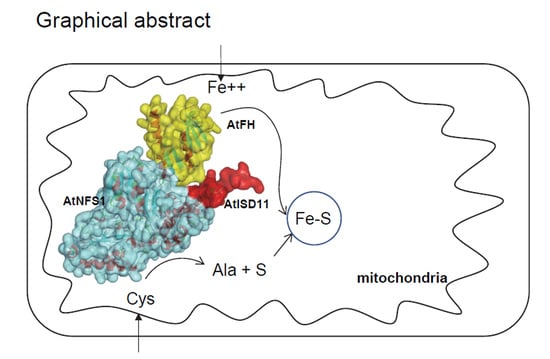
Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Effect of Frataxin (AtFH) and AtISD11 on Desulfurase (AtNFS1) Kinetic Parameters
2.2. Pull-Down Assays and Interaction Studies between AtNFS1, AtFH, and AtISD11
2.3. Evaluation of the Attenuation of Fenton Reaction in the Presence of AtFH, AtISD11, and AtNFS1
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression and Purification of AtNFS1, AtFH, and AtISD11
4.2. Cysteine Desulfurase Assay
4.3. Pull-Down Assays
4.4. Oxidative Degradation Assays
4.5. Additional Methods
4.6. Statistical Analyses
Author Contributions
Funding
Conflicts of Interest
References
- Couturier, J.; Touraine, B.; Briat, J.-F.; Gaymard, F.; Rouhier, N. The iron-sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front. Plant Sci. 2013, 4, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.M.; Møller, S.G. Iron–sulfur clusters: Biogenesis, molecular mechanisms, and their functional significance. Antioxid. Redox Signal. 2011, 15, 271–307. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.C.; Dean, D.R.; Hu, Y.; Ribbe, M.W. Formation and insertion of the nitrogenase iron-molybdenum cofactor. Chem. Rev. 2004, 104, 1159–1173. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.M.; Ludden, P.W. Maturation of nitrogenase: A biochemical puzzle. J. Bacteriol. 2005, 187, 405–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzer, S.I.; Hantke, K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 1999, 181, 3307–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Tokumoto, U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 2002, 277, 28380–28383. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Solodovnikova, N.; Nicholson, T.; Antholine, W.; Walden, W.E. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 2003, 22, 4826–4835. [Google Scholar] [CrossRef] [Green Version]
- Balk, J.; Lobreaux, S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005, 10, 324–331. [Google Scholar] [CrossRef]
- Balk, J.; Pilon, M. Ancient and essential: The assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011, 16, 218–226. [Google Scholar] [CrossRef]
- Lill, R. Function and biogenesis of iron–sulphur proteins. Nature 2009, 460, 831. [Google Scholar] [CrossRef]
- Lill, R.; Mühlenhoff, U. Iron–sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 2005, 30, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihara, H.; Kurihara, T.; Yoshimura, T.; Soda, K.; Esaki, N. Cysteine Sulfinate Desulfinase, a NIFS-like Protein ofEscherichia coli with Selenocysteine Lyase and Cysteine Desulfurase Activities gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 1997, 272, 22417–22424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; White, R.H.; Cash, V.L.; Jack, R.F.; Dean, D.R. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 1993, 90, 2754–2758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazzon, A.P.; Ramirez, M.V.; Warek, U.; Balk, J.; Frazzon, J.; Dean, D.R.; Winkel, B.S. Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol. Biol. 2007, 64, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Kushnir, S.; Babiychuk, E.; Storozhenko, S.; Davey, M.W.; Papenbrock, J.; De Rycke, R.; Engler, G.; Stephan, U.W.; Lange, H.; Kispal, G.; et al. A Mutation of the Mitochondrial ABC Transporter Sta1 Leads to Dwarfism and Chlorosis in the Arabidopsis Mutant starik. Plant Cell 2001, 13, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Pilon-Smits, E.A.; Garifullina, G.F.; Abdel-Ghany, S.; Kato, S.; Mihara, H.; Hale, K.L.; Burkhead, J.L.; Esaki, N.; Kurihara, T.; Pilon, M. Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol. 2002, 130, 1309–1318. [Google Scholar] [CrossRef] [Green Version]
- Sébastien, L.; Touraine, B.; Briat, J.-F.; Lobréaux, S. The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem. J. 2002, 366, 557–564. [Google Scholar]
- Turowski, V.R.; Busi, M.V.; Gomez-Casati, D.F. Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana. Mol. Plant 2012, 5, 1001–1010. [Google Scholar] [CrossRef] [Green Version]
- Busi, M.V.; Gomez-Casati, D.F. Exploring frataxin function. IUBMB Life 2012, 64, 56–63. [Google Scholar] [CrossRef]
- Castro, I.H.; Pignataro, M.F.; Sewell, K.E.; Espeche, L.D.; Herrera, M.G.; Noguera, M.E.; Dain, L.; Nadra, A.D.; Aran, M.; Smal, C.; et al. Frataxin Structure and Function. Sub-Cell. Biochem. 2019, 93, 393–438. [Google Scholar] [CrossRef]
- Babcock, M.; de Silva, D.; Oaks, R.; Davis-Kaplan, S.; Jiralerspong, S.; Montermini, L.; Pandolfo, M.; Kaplan, J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 1997, 276, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Lesuisse, E.; Santos, R.; Matzanke, B.F.; Knight, S.A.; Camadro, J.M.; Dancis, A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1). Human Mol. Genet. 2003, 12, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Chen, O.S.; Hemenway, S.; Kaplan, J. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: Evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12321–12326. [Google Scholar] [CrossRef] [Green Version]
- Busi, M.V.; Maliandi, M.V.; Valdez, H.; Clemente, M.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe–S proteins and induces oxidative stress. Plant J. 2006, 48, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Ristow, M.; Pfister, M.F.; Yee, A.J.; Schubert, M.; Michael, L.; Zhang, C.Y.; Ueki, K.; Michael, M.D., II; Lowell, B.B.; Kahn, C.R. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc. Natl. Acad. Sci. USA 2000, 97, 12239–12243. [Google Scholar] [CrossRef] [Green Version]
- Isaya, G.; O’Neill, H.A.; Gakh, O.; Park, S.; Mantcheva, R.; Mooney, S.M. Functional studies of frataxin. Acta Paediatr. 2004, 93, 68–71, discussion 72-63. [Google Scholar] [CrossRef]
- Gervason, S.; Larkem, D.; Mansour, A.B.; Botzanowski, T.; Muller, C.S.; Pecqueur, L.; Le Pavec, G.; Delaunay-Moisan, A.; Brun, O.; Agramunt, J.; et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 2019, 10, 3566. [Google Scholar] [CrossRef] [Green Version]
- Busi, M.V.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Functional and molecular characterization of the frataxin homolog from Arabidopsis thaliana. FEBS Lett. 2004, 576, 141–144. [Google Scholar] [CrossRef] [Green Version]
- Buchensky, C.; Sanchez, M.; Carrillo, M.; Palacios, O.; Capdevila, M.; Dominguez-Vera, J.M.; Busi, M.V.; Atrian, S.; Pagani, M.A.; Gomez-Casati, D.F. Identification of two frataxin isoforms in Zea mays: Structural and functional studies. Biochimie 2017, 140, 34–47. [Google Scholar] [CrossRef]
- Sanchez, M.; Palacios, O.; Buchensky, C.; Sabio, L.; Gomez-Casati, D.F.; Pagani, M.A.; Capdevila, M.; Atrian, S.; Dominguez-Vera, J.M. Copper redox chemistry of plant frataxins. J. Inorg. Biochem. 2018, 180, 135–140. [Google Scholar] [CrossRef]
- Maliandi, M.V.; Busi, M.V.; Clemente, M.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Expression and one-step purification of recombinant Arabidopsis thaliana frataxin homolog (AtFH). Protein Expr. Purif. 2007, 51, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Turowski, V.R.; Aknin, C.; Maliandi, M.V.; Buchensky, C.; Leaden, L.; Peralta, D.A.; Busi, M.V.; Araya, A.; Gomez-Casati, D.F. Frataxin Is Localized to Both the Chloroplast and Mitochondrion and Is Involved in Chloroplast Fe-S Protein Function in Arabidopsis. PLoS ONE 2015, 10, e0141443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Casati, D.F.; Busi, M.V.; Pagani, M.A. Plant Frataxin in Metal Metabolism. Front. Plant Sci. 2018, 9, 1706. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Mühlenhoff, U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008, 77, 669–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noguera, M.E.; Roman, E.A.; Rigal, J.B.; Cousido-Siah, A.; Mitschler, A.; Podjarny, A.; Santos, J. Structural characterization of metal binding to a cold-adapted frataxin. J. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2015, 20, 653–664. [Google Scholar] [CrossRef]
- Armas, A.M.; Balparda, M.; Terenzi, A.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Ferrochelatase activity of plant frataxin. Biochimie 2019, 156, 118–122. [Google Scholar] [CrossRef]
- Patra, S.; Barondeau, D.P. Mechanism of activation of the human cysteine desulfurase complex by frataxin. Proc. Natl. Acad.Sci. USA 2019, 116, 19421–19430. [Google Scholar] [CrossRef] [Green Version]
- Adam, A.C.; Bornhovd, C.; Prokisch, H.; Neupert, W.; Hell, K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006, 25, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Friemel, M.; Marelja, Z.; Li, K.; Leimkuhler, S. The N-Terminus of Iron-Sulfur Cluster Assembly Factor ISD11 Is Crucial for Subcellular Targeting and Interaction with l-Cysteine Desulfurase NFS1. Biochemistry 2017, 56, 1797–1808. [Google Scholar] [CrossRef]
- Atkinson, A.; Smith, P.; Fox, J.L.; Cui, T.Z.; Khalimonchuk, O.; Winge, D.R. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell. Biol. 2011, 31, 3988–3996. [Google Scholar] [CrossRef] [Green Version]
- Richards, T.A.; van der Giezen, M. Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol Evol 2006, 23, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Napoli, E.; Cortopassi, G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Human Mol. Genet. 2007, 16, 929–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cory, S.A.; Van Vranken, J.G.; Brignole, E.J.; Patra, S.; Winge, D.R.; Drennan, C.L.; Rutter, J.; Barondeau, D.P. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. USA 2017, 114, E5325–E5334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boniecki, M.T.; Freibert, S.A.; Muhlenhoff, U.; Lill, R.; Cygler, M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 2017, 8, 1287. [Google Scholar] [CrossRef] [Green Version]
- Wiedemann, N.; Urzica, E.; Guiard, B.; Muller, H.; Lohaus, C.; Meyer, H.E.; Ryan, M.T.; Meisinger, C.; Muhlenhoff, U.; Lill, R.; et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006, 25, 184–195. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, P.P.; Srivastava, S.; Kumar, S.K.P.; Sinha, D.; D’Silva, P. Mapping Key Residues of ISD11 Critical for NFS1-ISD11 Subcomplex Stability: IMPLICATIONS IN THE DEVELOPMENT OF MITOCHONDRIAL DISORDER, COXPD19. J. Biol. Chem. 2015, 290, 25876–25890. [Google Scholar] [CrossRef] [Green Version]
- Fox, N.G.; Yu, X.; Feng, X.; Bailey, H.J.; Martelli, A.; Nabhan, J.F.; Strain-Damerell, C.; Bulawa, C.; Yue, W.W.; Han, S. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 2019, 10, 2210. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Gakh, O.; Mooney, S.M.; Isaya, G. The ferroxidase activity of yeast frataxin. J. Biol. Chem. 2002, 277, 38589–38595. [Google Scholar] [CrossRef] [Green Version]
- Mühlenhoff, U.; Richhardt, N.; Ristow, M.; Kispal, G.; Lill, R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Human Mol. Genet. 2002, 11, 2025–2036. [Google Scholar] [CrossRef] [Green Version]
- Bencze, K.Z.; Kondapalli, K.C.; Cook, J.D.; McMahon, S.; Millán-Pacheco, C.; Pastor, N.; Stemmler, T.L. The structure and function of frataxin. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 269–291. [Google Scholar] [CrossRef] [Green Version]
- Dhe-Paganon, S.; Shigeta, R.; Chi, Y.I.; Ristow, M.; Shoelson, S.E. Crystal structure of human frataxin. J. Biol. Chem. 2000, 275, 30753–30756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neet, K.E.; Ainslie, G.R., Jr. Hysteretic enzymes. Methods Enzymol. 1980, 64, 192–226. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Fox, N.G.; Tsai, C.L.; Winn, A.M.; Barondeau, D.P. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 2014, 53, 4904–4913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, A.; Gordon, D.M.; Pain, J.; Stemmler, T.L.; Dancis, A.; Pain, D. Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J. Biol. Chem. 2013, 288, 36773–36786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, A.; Elduque, X.; Cornu, D.; Belot, L.; Le Caer, J.P.; Grandas, A.; Toledano, M.B.; D’Autreaux, B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 2015, 6, 5686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.L.; Barondeau, D.P. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 2010, 49, 9132–9139. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Meisel, J.D.; Patra, S.; Wang, H.; Grange, R.M.H.; Kim, S.H.; Calvo, S.E.; Orefice, L.L.; Nagashima, F.; Ichinose, F.; et al. Hypoxia Rescues Frataxin Loss by Restoring Iron Sulfur Cluster Biogenesis. Cell 2019, 177, 1507–1521.e1516. [Google Scholar] [CrossRef]
- Napoli, E.; Morin, D.; Bernhardt, R.; Buckpitt, A.; Cortopassi, G. Hemin rescues adrenodoxin, heme a and cytochrome oxidase activity in frataxin-deficient oligodendroglioma cells. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2007, 1772, 773–780. [Google Scholar] [CrossRef] [Green Version]
- Maliandi, M.V.; Busi, M.V.; Turowski, V.R.; Leaden, L.; Araya, A.; Gomez-Casati, D.F. The mitochondrial protein frataxin is essential for heme biosynthesis in plants. FEBS J. 2011, 278, 470–481. [Google Scholar] [CrossRef]
- Cai, K.; Frederick, R.O.; Dashti, H.; Markley, J.L. Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering. Structure 2018, 26, 1127–1136.e1124. [Google Scholar] [CrossRef] [Green Version]
- Foury, F.; Cazzalini, O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997, 411, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Lin, G.; Haelterman, N.A.; Ho, T.S.; Li, T.; Li, Z.; Duraine, L.; Graham, B.H.; Jaiswal, M.; Yamamoto, S.; et al. Loss of Frataxin induces iron toxicity, sphingolipid synthesis, and Pdk1/Mef2 activation, leading to neurodegeneration. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.; Colman, M.J.; Gomez-Casati, D.F.; Lamattina, L.; Zabaleta, E.J. Nitric oxide accumulation is required to protect against iron-mediated oxidative stress in frataxin-deficient Arabidopsis plants. FEBS Lett. 2009, 583, 542–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, J.L.; Blake, J.C.; Chamberlain, S.; Thomas, P.K.; Cooper, J.M.; Schapira, A.H. Clinical, biochemical and molecular genetic correlations in Friedreich’s ataxia. Human Mol. Genet. 2000, 9, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Bonilha da Silva, C.; Bergo, F.P.G.; D’Abreu, A.; Cendes, F.; Lopes-Cendes, I.; Franca, M.C., Jr. Dentate nuclei T2 relaxometry is a reliable neuroimaging marker in Friedreich’s ataxia. Eur. J. Neurol. 2014, 21, 1131–1136. [Google Scholar] [CrossRef]
- Armas, A.M.; Balparda, M.; Turowski, V.R.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Altered levels of mitochondrial NFS1 affect cellular Fe and S contents in plants. Plant. Cell Rep. 2019, 38, 981–990. [Google Scholar] [CrossRef]
- Moore, S.; Stein, W.H. Photometric nin-hydrin method for use in the ehromatography of amino acids. J. Biol. Chem. 1948, 176, 367–388. [Google Scholar]
- Sheng, S.; Kraft, J.J.; Schuster, S.M. A specific quantitative colorimetric assay for L-asparagine. Anal. Biochem. 1993, 211, 242–249. [Google Scholar] [CrossRef]
- Brooks, S.P. A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques 1992, 13, 906–911. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pons, J.L.; Labesse, G. @TOME-2: A new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009, 37, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PyMol by Schrödinger. The PyMOL Molecular Graphics System. Available online: https://pymol.org/2/ (accessed on 21 July 2020).
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armas, A.M.; Balparda, M.; Terenzi, A.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana. Plants 2020, 9, 1171. https://doi.org/10.3390/plants9091171
Armas AM, Balparda M, Terenzi A, Busi MV, Pagani MA, Gomez-Casati DF. Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana. Plants. 2020; 9(9):1171. https://doi.org/10.3390/plants9091171
Chicago/Turabian StyleArmas, Alejandro M., Manuel Balparda, Agustina Terenzi, Maria V. Busi, Maria A. Pagani, and Diego F. Gomez-Casati. 2020. "Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana" Plants 9, no. 9: 1171. https://doi.org/10.3390/plants9091171