Jasmonate Signalling Contributes to Primary Root Inhibition Upon Oxygen Deficiency in Arabidopsis thaliana
Abstract
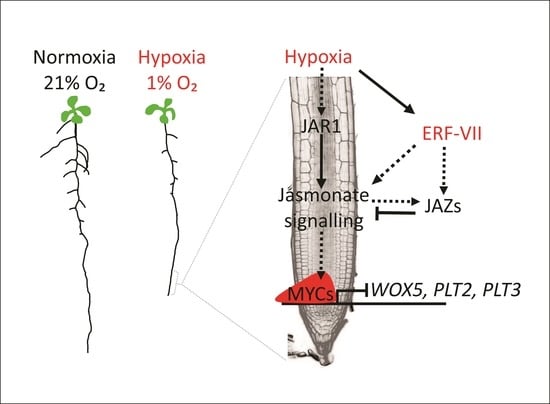
:1. Introduction
2. Results
2.1. Primary Root Growth Is Transiently Inhibited under Hypoxia
2.2. Root Meristem Regulators Are Repressed by Hypoxia
2.3. Assessment of Jasmonate Levels and Activity in Hypoxic Roots
2.4. JA-Responsive Genes Are Affected in Hypoxic Roots
2.5. JA Signaling Is Active under Hypoxia and Contributes to Root Growth Restriction
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Measurement of Primary Root Length
4.3. Confocal Imaging
4.4. In Silico Analysis of Differentially Expressed Genes
4.5. RNA Extraction and Gene Expression Analysis by Real-Time qRT-PCR
4.6. Hormone Quantification
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Akgerman, A.; Gainer, J.L. Diffusion of Gases in Liquids. Ind. Eng. Chem. Fundam. 1972. [Google Scholar] [CrossRef]
- Gibbs, J.; Greenway, H. Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Fagerstedt, K.V. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol. Biochem. 2010, 48, 359–373. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.T.; Licausi, F. Oxygen Sensing and Signaling. Annu. Rev. Plant Biol. 2014, 66, 150112150216002. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: an overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Mendiondo, G.M.; Gibbs, D.J.; Szurman-Zubrzycka, M.; Korn, A.; Marquez, J.; Szarejko, I.; Maluszynski, M.; King, J.; Axcell, B.; Smart, K.; et al. Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol. J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cukrov, D.; Zermiani, M.; Brizzolara, S.; Cestaro, A.; Licausi, F.; Luchinat, C.; Santucci, C.; Tenori, L.; Van Veen, H.; Zuccolo, A.; et al. Extreme Hypoxic Conditions Induce Selective Molecular Responses and Metabolic Reset in Detached Apple Fruit. Front. Plant Sci. 2016. [Google Scholar] [CrossRef] [Green Version]
- Bui, L.T.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Licausi, F. Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 2015, 236, 37–43. [Google Scholar] [CrossRef]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII transcription factors bind an evolutionarily-conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 2015, TPC2015-00866-RA. [Google Scholar] [CrossRef] [Green Version]
- Kosmacz, M.; Parlanti, S.; Schwarzländer, M.; Kragler, F.; Licausi, F.; Van Dongen, J.T. The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant, Cell Environ. 2015, 38, 1094–1103. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Lee, S.C.; Md Isa, N.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.-M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.D.; Klecker, M.; Hopkinson, R.J.; Weits, D.A.; Mueller, C.; Naumann, C.; O’Neill, R.; Wickens, J.; Yang, J.; Brooks-Bartlett, J.C.; et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- White, M.D.; Kamps, J.J.A.G.; East, S.; Taylor Kearney, L.J.; Flashman, E. The plant cysteine oxidases from Arabidopsis thaliana are kinetically tailored to act as oxygen sensors. J. Biol. Chem. 2018, 293, 11786–11795. [Google Scholar] [CrossRef] [Green Version]
- Garzón, M.; Eifler, K.; Faust, A.; Scheel, H.; Hofmann, K.; Koncz, C.; Yephremov, A.; Bachmair, A. PRT6 /At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 2007, 581, 3189–3196. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.V.; Iyer, S.; Amerhauser, C.; Lehmann, M.; van Dongen, J.T.; Geigenberger, P. RAP2.12 oxygen sensing regulates plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 2016, 172, 00460. [Google Scholar] [CrossRef] [Green Version]
- Giuntoli, B.; Shukla, V.; Maggiorelli, F.; Giorgi, F.M.; Lombardi, L.; Perata, P.; Licausi, F. Age-dependent regulation of ERF-VII transcription factor activity in Arabidopsis thaliana. Plant. Cell Environ. 2017. [Google Scholar] [CrossRef]
- Wright, A.J.; de Kroon, H.; Visser, E.J.W.; Buchmann, T.; Ebeling, A.; Eisenhauer, N.; Fischer, C.; Hildebrandt, A.; Ravenek, J.; Roscher, C.; et al. Plants are less negatively affected by flooding when growing in species-rich plant communities. New Phytol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Jitsuyama, Y. Morphological root responses of soybean to rhizosphere hypoxia reflect waterlogging tolerance. Can. J. Plant Sci. 2015, 95, 999–1005. [Google Scholar] [CrossRef]
- Cardoso, J.A.; Jiménez, J.D.L.C.; Rao, I.M. Waterlogging-induced changes in root architecture of germplasm accessions of the tropical forage grass Brachiaria humidicola. AoB Plants 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzesiak, M.T.; Ostrowska, A.; Hura, K.; Rut, G.; Janowiak, F.; Rzepka, A.; Hura, T.; Grzesiak, S. Interspecific differences in root architecture among maize and triticale genotypes grown under drought, waterlogging and soil compaction. Acta Physiol. Plant. 2014. [Google Scholar] [CrossRef] [Green Version]
- Cornelious, B.; Chen, P.; Chen, Y.; De Leon, N.; Shannon, J.G.; Wang, D. Identification of QTLs underlying water-logging tolerance in soybean. Mol. Breed. 2005. [Google Scholar] [CrossRef]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant root growth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Lynch, J. Root Architecture and Plant Productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Visser, E.; Cohen, J.D.; Barendse, G.; Blom, C.; Voesenek, L. An Ethylene-Mediated Increase in Sensitivity to Auxin Induces Adventitious Root Formation in Flooded Rumex palustris Sm. Plant Physiol. 1996. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Kumar, P. Lysigenous aerenchyma formation involves non-apoptotic programmed cell death in rice (Oryza sativa L.) roots. Physiol. Mol. Biol. Plants 2012, 18, 1. [Google Scholar] [CrossRef] [Green Version]
- THOMAS, A.L.; GUERREIRO, S.M.C.; SODEK, L. Aerenchyma Formation and Recovery from Hypoxia of the Flooded Root System of Nodulated Soybean. Ann. Bot. 2005, 96, 1191. [Google Scholar] [CrossRef]
- van Dongen, J.T.; Fröhlich, A.; Ramírez-Aguilar, S.J.; Schauer, N.; Fernie, A.R.; Erban, A.; Kopka, J.; Clark, J.; Langer, A.; Geigenberger, P. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann. Bot. 2009, 103, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Zanetti, M.E.; Jang, C.J.H.; Holtan, H.E.; Repetti, P.P.; Galbraith, D.W.; Girke, T.; Bailey-Serres, J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. 2009, 106, 18843–18848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.B.; Fenning, T.M.; Jenkins, W. Aerenchyma (gas-space) formation in adventitious roots of rice (Oryza sativa L.) is not controlled by ethylene or small partial pressures of oxygen. J. Exp. Bot. 1985, 36, 1566–1572. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Rijnders, J.H.G.M.; Peeters, A.J.M.; Van De Steeg, H.M.; De Kroon, H. Plant hormones regulate fast shoot elongation under water: From genes to communities. Ecology 2004, 85, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Steffens, B.; Wang, J.; Sauter, M. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 2006, 223, 604–612. [Google Scholar] [CrossRef]
- Nishiuchi, S.; Yamauchi, T.; Takahashi, H.; Kotula, L.; Nakazono, M. Mechanisms for coping with submergence and waterlogging in rice. Rice 2012, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009. [Google Scholar] [CrossRef] [Green Version]
- McConn, M.; Browse, J. The Critical Requirement for Linolenic Acid Is Pollen Development, Not Photosynthesis, in an Arabidopsis Mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, 1991–2001. [Google Scholar] [CrossRef] [Green Version]
- Xie, D. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Montiel, G.; Zarei, A.; Körbes, A.P.; Memelink, J. The jasmonate-responsive element from the ORCA3 promoter from catharanthus roseus is active in arabidopsis and is controlled by the transcription factor AtMYC2. Plant Cell Physiol. 2011, 52, 578–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The Basic Helix-Loop-Helix Transcription Factor MYC2 Directly Represses PLETHORA Expression during Jasmonate-Mediated Modulation of the Root Stem Cell Niche in Arabidopsis. Plant Cell Online 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dathe, W.; Rönsch, H.; Preiss, A.; Schade, W.; Sembdner, G.; Schreiber, K. Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 1981, 153, 530–535. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Poirier, Y.; Antonenkov, V.D.; Glumoff, T.; Hiltunen, J.K. Peroxisomal β-oxidation—A metabolic pathway with multiple functions. Biochim. Biophys. Acta - Mol. Cell Res. 2006, 1763, 1413–1426. [Google Scholar] [CrossRef] [Green Version]
- Abbas, M.; Berckhan, S.; Rooney, D.J.; Gibbs, D.J.; Vicente Conde, J.; Sousa Correia, C.; Bassel, G.W.; Marín-De La Rosa, N.; León, J.; Alabadí, D.; et al. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 2015, 25, 1483–1488. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inzé, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinformatics 2008, 2008, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Goossens, A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eysholdt-Derzso, E.; Sauter, M. Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol. 2017, 175, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Yazdanbakhsh, N.; Fisahn, J. Analysis of Arabidopsis thaliana root growth kinetics with high temporal and spatial resolution. Ann. Bot. 2010. [Google Scholar] [CrossRef] [PubMed]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the enzymatic activity in the lipoxygenase gene family of arabidopsis thaliana. Lipids 2009, 44, 85–95. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Martinoia, E.; Farmer, E.E. Emerging Jasmonate Transporters. Mol. Plant 2017. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, J.; Li, S.; Huang, G.; Skilling, S.J.; Wang, L.; Li, L.; Li, M.; Yuan, L.; Liu, P. Transporter-Mediated Nuclear Entry of Jasmonoyl-Isoleucine Is Essential for Jasmonate Signaling. Mol. Plant 2017. [Google Scholar] [CrossRef] [Green Version]
- de Marchi, R.; Sorel, M.; Mooney, B.; Fudal, I.; Goslin, K.; Kwaśniewska, K.; Ryan, P.T.; Pfalz, M.; Kroymann, J.; Pollmann, S.; et al. The N-end rule pathway regulates pathogen responses in plants. Sci. Rep. 2016, 6, 26020. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A Trihelix DNA Binding Protein Counterbalances Hypoxia-Responsive Transcriptional Activation in Arabidopsis. PLoS Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis Basic Helix-Loop-Helix Transcription Factors MYC2, MYC3, and MYC4 Regulate Glucosinolate Biosynthesis, Insect Performance, and Feeding Behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mähönen, A.P.; Ten Tusscher, K.; Siligato, R.; Smetana, O.; Díaz-Triviño, S.; Salojärvi, J.; Wachsman, G.; Prasad, K.; Heidstra, R.; Scheres, B. PLETHORA gradient formation mechanism separates auxin responses. Nature 2014, 515, 125–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrieu, A.; Champion, A.; Legrand, J.; Lavenus, J.; Mast, D.; Brunoud, G.; Oh, J.; Guyomarc’h, S.; Pizot, M.; Farmer, E.E.; et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Commun. 2015, 6, 6043. [Google Scholar] [CrossRef] [Green Version]
- Armengaud, P. EZ-Rhizo software. Plant Signal. Behav. 2009, 4, 139–141. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Inzé, D.; Goossens, A. Jasmonate-inducible gene: what does it mean? Trends Plant Sci. 2009, 14, 87–91. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous hypoxia in lateral root primordia controls root architecture by antagonizing auxin signaling in Arabidopsis. Mol. Plant 2019. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Floková, K.; Tarkowská, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novák, O. UHPLC–MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, V.; Lombardi, L.; Pencik, A.; Novak, O.; Weits, D.A.; Loreti, E.; Perata, P.; Giuntoli, B.; Licausi, F. Jasmonate Signalling Contributes to Primary Root Inhibition Upon Oxygen Deficiency in Arabidopsis thaliana. Plants 2020, 9, 1046. https://doi.org/10.3390/plants9081046
Shukla V, Lombardi L, Pencik A, Novak O, Weits DA, Loreti E, Perata P, Giuntoli B, Licausi F. Jasmonate Signalling Contributes to Primary Root Inhibition Upon Oxygen Deficiency in Arabidopsis thaliana. Plants. 2020; 9(8):1046. https://doi.org/10.3390/plants9081046
Chicago/Turabian StyleShukla, Vinay, Lara Lombardi, Ales Pencik, Ondrej Novak, Daan A. Weits, Elena Loreti, Pierdomenico Perata, Beatrice Giuntoli, and Francesco Licausi. 2020. "Jasmonate Signalling Contributes to Primary Root Inhibition Upon Oxygen Deficiency in Arabidopsis thaliana" Plants 9, no. 8: 1046. https://doi.org/10.3390/plants9081046