Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle
Abstract
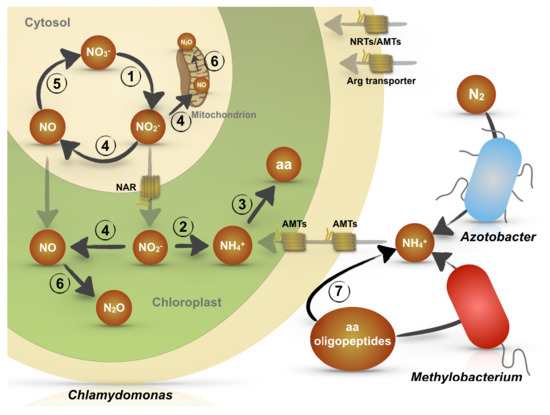
:1. Introduction
2. The Role of Chlamydomonas reinhardtii in the N Cycle
2.1. Ammonification and Mutualism with Bacteria for N Scavenging from Amino Acids and Peptides
2.2. Nitrate Denitrification
2.2.1. The NO3−/NO3− Cycle
2.2.2. NO Synthesis and Reduction: A Prokaryote Pathway
3. N2O Production and Enzymes in Other Microalgae
4. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Stein, L.Y.; Klotz, M.G. The nitrogen cycle. Curr. Biol. 2016, 26, R94–R98. [Google Scholar] [CrossRef] [Green Version]
- Zerkle, A.L.; Mikhail, S. The geobiological nitrogen cycle: From microbes to the mantle. Geobiology 2017, 15, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Burris, R.H.; Roberts, G.P. Biological nitrogen fixation. Annu. Rev. Nutr. 1993, 13, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Harding, K.; Turk-Kubo, K.A.; Sipler, R.E.; Mills, M.M.; Bronk, D.A.; Zehr, J.P. Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. Proc. Natl. Acad. Sci. USA 2018, 115, 13371–13375. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Marin, M.D.C.; Shilova, I.N.; Shi, T.; Farnelid, H.; Cabello, A.M.; Zehr, J.P. The transcriptional cycle is suited to daytime N2 fixation in the unicellular cyanobacterium Candidatus Atelocyanobacterium thalassa (UCYN-A). mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Cheng, X.; Wang, Q. Enhanced lipid production in Chlamydomonas reinhardtii by co-culturing with Azotobacter chroococcum. Front. Plant Sci. 2018, 9, 741. [Google Scholar] [CrossRef]
- Erisman, J.Z.; Sutton, M.A.; Galloway, J.; Klimont, Z.; Winiwarter, W. How a century of ammonia synthesis changed the world. Nat. Geosci. 2008, 1, 636–639. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Ravishankara, A.R.; Daniel, J.S.; Portmann, R.W. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 2009, 326, 123–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crutzen, P.J.; Moiser, A.R.; Smith, K.A.; Winiwarter, W. N2O release from agro-biofuel production negates global warming reduction by replacing fossil fuels. Atmos. Chem. Phys. 2008, 8, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Davidson, A. The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nat. Geosci. 2009, 2, 659–662. [Google Scholar] [CrossRef]
- Kamp, A.; Stief, P.; Knappe, J.; de Beer, D. Response of the ubiquitous pelagic diatom Thalassiosira weissflogii to darkness and anoxia. PLoS ONE 2013, 8, e82605. [Google Scholar] [CrossRef] [PubMed]
- Kamp, A.; de Beer, D.; Nitsch, J.L.; Lavik, G.; Stief, P. Diatoms respire nitrate to survive dark and anoxic conditions. Proc. Natl. Acad. Sci. USA 2011, 108, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risgaard-Petersen, N.; Langezaal, A.M.; Ingvardsen, S.; Schmid, M.C.; Jetten, M.S.; Op den Camp, H.J.; Derksen, J.W.; Pina-Ochoa, E.; Eriksson, S.P.; Nielsen, L.P.; et al. Evidence for complete denitrification in a benthic foraminifer. Nature 2006, 443, 93–96. [Google Scholar] [CrossRef]
- Piña-Ochoa, E.; Hogslund, S.; Geslin, E.; Cedhagen, T.; Revsbech, N.P.; Nielsen, L.P.; Schweizer, M.; Jorissen, F.; Rysgaard, S.; Risgaard-Petersen, N. Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc. Natl. Acad. Sci. USA 2010, 107, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- Bernhard, J.M.; Casciotti, K.L.; McIlvin, M.R.; Beaudoin, D.J.; Visscher, P.T.; Edgcomb, V.P. Potential importance of physiologically diverse benthic foraminifera in sedimentary nitrate storage and respiration. J. Geophys. Res. 2012, 117, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kamp, A.; Hogslund, S.; Risgaard-Petersen, N.; Stief, P. Nitrate storage and dissimilatory nitrate reduction by eukaryotic microbes. Front. Microbiol. 2015, 6, 1492. [Google Scholar] [CrossRef] [Green Version]
- Woehle, C.; Roy, A.S.; Glock, N.; Wein, T.; Weissenbach, J.; Rosenstiel, P.; Hiebenthal, C.; Michels, J.; Schonfeld, J.; Dagan, T. A Novel eukaryotic denitrification pathway in foraminifera. Curr. Biol. 2018, 28, 2536–2543. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Fushinobu, S.; Zhou, S.; Wakagi, T.; Shoun, H. Eukaryotic nirK genes encoding copper-containing nitrite reductase: Originating from the protomitochondrion? Appl. Environ. Microbiol. 2009, 75, 2652–2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoun, H.; Fushinobu, S.; Jiang, L.; Kim, S.W.; Wakagi, T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1186–1194. [Google Scholar] [CrossRef] [Green Version]
- Maeda, K.; Spor, A.; Edel-Hermann, V.; Heraud, C.; Breuil, M.C.; Bizouard, F.; Toyoda, S.; Yoshida, N.; Steinberg, C.; Philippot, L. N2O production, a widespread trait in fungi. Sci. Rep. 2015, 5, 9697. [Google Scholar] [CrossRef] [Green Version]
- Higgins, S.A.; Welsh, A.; Orellana, L.H.; Konstantinidis, K.T.; Chee-Sanford, J.C.; Sanford, R.A.; Schadt, C.W.; Loffler, F.E. Detection and diversity of fungal nitric oxide reductase genes (p450nor) in agricultural soils. Appl. Environ. Microbiol. 2016, 82, 2919–2928. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Vine, C.E.; Balasiny, B.K.; Rizk, J.; Bradley, C.L.; Tinajero-Trejo, M.; Poole, R.K.; Bergaust, L.L.; Bakken, L.R.; Cole, J.A. The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol. Microbiol. 2016, 100, 877–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plouviez, M.; Wheeler, D.; Shilton, A.; Packer, M.A.; McLenachan, P.A.; Sanz-Luque, E.; Ocana-Calahorro, F.; Fernandez, E.; Guieysse, B. The biosynthesis of nitrous oxide in the green alga Chlamydomonas reinhardtii. Plant J. 2017, 91, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Burlacot, A.; Richaud, P.; Gosset, A.; Li-Beisson, Y.; Peltier, G. Algal photosynthesis converts nitric oxide into nitrous oxide. Proc. Natl. Acad. Sci. USA 2020, 117, 2704–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plouviez, M.; Shilton, A.; Packer, M.A.; Guieysse, B. Nitrous oxide emissions from microalgae: Potential pathways and significance. J. Appl. Phycol. 2019, 31, 1–8. [Google Scholar] [CrossRef]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. eLife 2018, 7. [Google Scholar] [CrossRef]
- Proschold, T.; Marin, B.; Schlosser, U.G.; Melkonian, M. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chlor´omonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 2001, 152, 265–300. [Google Scholar] [CrossRef]
- Salome, P.A.; Merchant, S.S. A series of fortunate events: Introducing Chlamydomonas as a reference organism. Plant Cell 2019, 31, 1682–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, E.; Galvan, A. Nitrate assimilation in Chlamydomonas. Eukaryot. Cell 2008, 7, 555–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Luque, E.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant. Sci. 2015, 6, 899. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, E.; Galvan, A. Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 2007, 58, 2279–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Ocana-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galvan, A.; Fernandez, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell. Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Düner, M.; Lambertz, J.; Mugge, C.; Hemschemeier, A. The soluble guanylate cyclase CYG12 is required for the acclimation to hypoxia and trophic regimes in Chlamydomonas reinhardtii. Plant J. 2018, 93, 311–337. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Luque, E.; Ocaña-Calahorro, F.; de Montaigu, A.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. THB1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity. Plant J. 2015, 81, 467–479. [Google Scholar] [CrossRef]
- Gerin, S.; Mathy, G.; Blomme, A.; Franck, F.; Sluse, F.E. Plasticity of the mitoproteome to nitrogen sources (nitrate and ammonium) in Chlamydomonas reinhardtii: The logic of Aox1 gene localization. Biochim. Biophys. Acta 2010, 1797, 994–1003. [Google Scholar] [CrossRef] [Green Version]
- van Lis, R.; Brugiere, S.; Baffert, C.; Coute, Y.; Nitschke, W.; Atteia, A. Hybrid cluster proteins in a photosynthetic microalga. FEBS J. 2020, 287, 721–735. [Google Scholar]
- Atteia, A.; Adrait, A.; Brugiere, S.; Tardif, M.; van Lis, R.; Deusch, O.; Dagan, T.; Kuhn, L.; Gontero, B.; Martin, W.; et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 2009, 26, 1533–1548. [Google Scholar] [CrossRef] [Green Version]
- Calatrava, V.; Hom, E.F.Y.; Llamas, A.; Fernandez, E.; Galvan, A. Nitrogen scavenging from amino acids and peptides in the model alga Chlamydomonas reinhardtii. The role of extracellular L-amino oxidase. Algal Res. 2019, 28, 101395. [Google Scholar] [CrossRef]
- Kirk, D.L.; Kirk, M.M. Carrier-mediated uptake of arginine and urea by Chlamydomonas reinhardtii. Plant Physiol. 1978, 61, 556–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Z.; Qingqing, R.; Chen, K.; Yang, L.; Cheng, Z.; Peng, K.; Zhu, Y.; Bai, Y.; Wang, Y. Study of amino acids as nitrogen source in Chlamydomonas reinhardtii. Phycol. Res. 2012, 60, 161–168. [Google Scholar] [CrossRef]
- Vallon, O.; Bulte, L.; Kuras, R.; Olive, J.; Wollman, F.A. Extensive accumulation of an extracellular L-amino-acid oxidase during gametogenesis of Chlamydomonas reinhardtii. Eur. J. Biochem. 1993, 215, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Schroda, M.; Vallon, O. Chaperones and Proteases. In The Chlamydomonas Sourcebook, 2nd ed.; Harris, E.H., Stern, D.B., Witman, G.B., Eds.; Academic Press: Oxford, UK, 2009; pp. 671–729. [Google Scholar]
- Schmollinger, S.; Muhlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Blanco, J.; Hidalgo-Martinez, J.; Cardenas, J. Extracellular deamination of L-amino acids by Chlamydomonas reinhardtii cells. Planta 1990, 182, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Kasai, K.; Ishikawa, T.; Nakamura, T.; Miura, T. Antibacterial properties of L-amino acid oxidase: Mechanisms of action and perspectives for therapeutic applications. Appl. Microbiol. Biotechnol. 2015, 99, 7847–7857. [Google Scholar] [CrossRef] [PubMed]
- Hunken, M.; Harder, J.; Kirst, G.O. Epiphytic bacteria on the Antarctic ice diatom Amphiprora kufferathii Manguin cleave hydrogen peroxide produced during algal photosynthesis. Plant Biol. 2008, 10, 519–526. [Google Scholar] [CrossRef]
- Calatrava, V.; Hom, E.F.Y.; Llamas, A.; Fernandez, E.; Galvan, A. OK, thanks! A new mutualism between Chlamydomonas and methylobacteria facilitates growth on amino acids and peptides. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant. Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Leon, J.; Costa-Broseta, A. Present knowledge and controversies, deficiencies, and misconceptions on nitric oxide synthesis, sensing, and signaling in plants. Plant Cell Environ. 2020, 43, 1–15. [Google Scholar] [CrossRef] [PubMed]
- de Montaigu, A.; Sanz-Luque, E.; Macias, M.I.; Galvan, A.; Fernandez, E. Transcriptional regulation of CDP1 and CYG56 is required for proper NH4+ sensing in Chlamydomonas. J. Exp. Bot. 2011, 62, 1425–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Sakihama, Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 2000, 468, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.J.; Stoimenova, M.; Kaiser, W.M. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 2005, 56, 2601–2609. [Google Scholar] [CrossRef]
- Jasid, S.; Simontacchi, M.; Bartoli, C.G.; Puntarulo, S. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 2006, 142, 1246–1255. [Google Scholar] [CrossRef] [Green Version]
- Tewari, R.K.; Prommer, J.; Watanabe, M. Endogenous nitric oxide generation in protoplast chloroplasts. Plant Cell Rep. 2013, 32, 31–44. [Google Scholar] [CrossRef]
- Corpas, F.J.; Del Rio, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019, 61, 803–816. [Google Scholar] [PubMed] [Green Version]
- Foresi, N.; Mayta, M.L.; Lodeyro, A.F.; Scuffi, D.; Correa-Aragunde, N.; Garcia-Mata, C.; Casalongue, C.; Carrillo, N.; Lamattina, L. Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis. Plant J. 2015, 82, 806–821. [Google Scholar] [CrossRef]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2. [Google Scholar] [CrossRef] [Green Version]
- del Rio, L.A.; Corpas, F.J.; Barroso, J.B. Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 2004, 65, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ballester, D.; Sanz-Luque, E.; Galvan, A.; Fernandez, E.; de Montaigu, A. Arginine is a component of the ammonium-CYG56 signalling cascade that represses genes of the nitrogen assimilation pathway in Chlamydomonas reinhardtii. PLoS ONE 2018, 13, e0196167. [Google Scholar] [CrossRef] [PubMed]
- Hemschemeier, A.; Duner, M.; Casero, D.; Merchant, S.S.; Winkler, M.; Happe, T. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc. Natl. Acad. Sci. USA 2013, 110, 10854–10859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folgosa, F.; Martins, M.C.; Teixeira, M. The multidomain flavodiiron protein from Clostridium difficile 630 is an NADH:oxygen oxidoreductase. Sci. Rep. 2018, 8, 10164. [Google Scholar] [CrossRef]
- Romao, C.V.; Vicente, J.B.; Borges, P.T.; Frazao, C.; Teixeira, M. The dual function of flavodiiron proteins: Oxygen and/or nitric oxide reductases. J. Biol. Inorg. Chem. 2016, 21, 39–52. [Google Scholar] [CrossRef]
- Seedorf, H.; Dreisbach, A.; Hedderich, R.; Shima, S.; Thauer, R.K. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 2004, 182, 126–137. [Google Scholar] [CrossRef]
- Smutna, T.; Goncalves, V.L.; Saraiva, L.M.; Tachezy, J.; Teixeira, M.; Hrdy, I. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: The terminal oxygen reductase. Eukaryot. Cell 2009, 8, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Di Matteo, A.; Scandurra, F.M.; Testa, F.; Forte, E.; Sarti, P.; Brunori, M.; Giuffre, A. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 2008, 283, 4061–4068. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.B.; Gomes, C.M.; Wasserfallen, A.; Teixeira, M. Module fusion in an A-type flavoprotein from the cyanobacterium Synechocystis condenses a multiple-component pathway in a single polypeptide chain. Biochem. Biophys. Res. Commun. 2002, 294, 82–87. [Google Scholar] [CrossRef]
- Silaghi-Dumitrescu, R.; Coulter, E.D.; Das, A.; Ljungdahl, L.G.; Jameson, G.N.; Huynh, B.H.; Kurtz, D.M., Jr. A flavodiiron protein and high molecular weight rubredoxin from Moorella thermoacetica with nitric oxide reductase activity. Biochemistry 2003, 42, 2806–2815. [Google Scholar] [CrossRef]
- Rodrigues, R.; Vicente, J.B.; Felix, R.; Oliveira, S.; Teixeira, M.; Rodrigues-Pousada, C. Desulfovibrio gigas flavodiiron protein affords protection against nitrosative stress in vivo. J. Bacteriol. 2006, 188, 2745–2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justino, M.C.; Vicente, J.B.; Teixeira, M.; Saraiva, L.M. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 2005, 280, 2636–2643. [Google Scholar] [CrossRef] [Green Version]
- Gardner, A.M.; Helmick, R.A.; Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002, 277, 8172–8177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaux, F.; Burlacot, A.; Mekhalfi, M.; Auroy, P.; Blangy, S.; Richaud, P.; Peltier, G. Flavodiiron proteins promote fast and transient O2 photoreduction in Chlamydomonas. Plant Physiol. 2017, 174, 1825–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mus, F.; Dubini, A.; Seibert, M.; Posewitz, M.C.; Grossman, A.R. Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 2007, 282, 25475–25486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, A.C.; Carter, C.J. The involvement of hybrid cluster protein 4, HCP4, in anaerobic metabolism in Chlamydomonas reinhardtii. PLoS ONE 2016, 11, e0149816. [Google Scholar] [CrossRef] [Green Version]
- Overeijnder, M.L.; Hagen, W.R.; Hagedoorn, P.L. A thermostable hybrid cluster protein from Pyrococcus furiosus: Effects of the loss of a three helix bundle subdomain. J. Biol. Inorg. Chem. 2009, 14, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Pesesky, M.; Zhang, L.; Huang, J.; Winkler, M.; Chistoserdova, L. A Complex interplay between nitric oxide, quorum sensing, and the unique secondary metabolite tundrenone constitutes the hypoxia response in Methylobacter. mSystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Seth, D.; Hess, D.T.; Hausladen, A.; Wang, L.; Wang, Y.J.; Stamler, J.S. A Multiplex enzymatic machinery for cellular protein s-nitrosylation. Mol. Cell 2018, 69, 451–464LO. [Google Scholar] [CrossRef] [Green Version]
- Bange, H.B.; Arevalo-Martiniez, D.L.; Paz, M.; Farias, L.; Kaiser, J.; Kock, A.; Law, C.S.; Rees, A.P.; Rehder, G.; Tortell, P.D.; et al. A harmonized nitrous oxide (N2O) ocean observation network for the 21st century. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Cohen, Y.; Gordon, L.G. Nitrous oxide in the oxygen minimum of the eastern tropical North Pacific: Evidence for its consumption during denitrification and possible mechanisms for its production. Deep Sea Res. 1978, 25, 509–524. [Google Scholar] [CrossRef]
- Smith, C.J.; DeLaune, R.D.; Patrick, W.H. Nitrous oxide emission from Gulf Coast wetlands. Geochim. Cosmochin. 1983, 47, 1805–1814. [Google Scholar] [CrossRef]
- Mengis, M.; Gächter, R.; Wehrli, B. Sources and sinks of nitrous oxide (N2O) in deep lakes. Biogeochemistry 1997, 38, 281–301. [Google Scholar] [CrossRef]
- Guieysse, B.; Plouviez, M.; Coilhac, M.; Cazali, L. Nitrous Oxide (N2O) production in axenic Chlorella vulgaris microalgae cultures: Evidence, putative pathways, and potential environmental impacts. Biogeosciences 2013, 10, 6737–6746. [Google Scholar] [CrossRef] [Green Version]
- Smeets, E.M.; Bouwman, L.F.; Stehfest, E.; Vuuren, D.P.; Posthuma, A. Contribution of N2O to the greenhouse gas balance of first-generation biofuels. Glob. Chang. Biol. 2009, 15, 1–23. [Google Scholar] [CrossRef]
- Bauer, S.K.; Grotz, L.S.; Connelly, E.B.; Colosi, L.M. Reevaluation of the global warming impacts of algae-derived biofuels to account for possible contributions of nitrous oxide. Bioresour. Technol. 2016, 218, 196–201. [Google Scholar] [CrossRef]
- Higgins, S.A.; Schadt, C.W.; Matheny, P.B.; Loffler, F.E. Phylogenomics reveal the dynamic evolution of fungal nitric oxide reductases and their relationship to secondary metabolism. Genome Biol. Evol. 2018, 10, 2474–2489. [Google Scholar] [CrossRef] [Green Version]
- Weather, P.J. N2O Evolution by green algae. Appl. Environ. Microbiol. 1984, 48, 1251–1253. [Google Scholar] [CrossRef] [Green Version]
- Florez-Leiva, L.; Tarifeño, E.; Cornejo, M.; Kiene, R.; Farias, L. High production of nitrous oxide (N2O), methane (CH4) and dimethylsulphoniopropionate (DMSP) in a massive marine phytoplankton culture. Biogeosci. Discuss. 2010, 7, 6705–6723. [Google Scholar] [CrossRef]
- Fagerstone, K.D.; Quinn, J.C.; Bradley, T.H.; De Long, S.K.; Marchese, A.J. Quantitative measurement of direct nitrous oxide emissions from microalgae cultivation. Environ. Sci. Technol. 2011, 45, 9449–9456. [Google Scholar] [CrossRef] [PubMed]

| Step | Enzyme | Gene ID | Regulation | Localization | Ref. | |
|---|---|---|---|---|---|---|
| 1 | NO3– + 2e– + 2H+→ NO2– + H2O | NR | Cre09.g410950 | NO3– | Cytosol | [33] |
| 2 | NO2– + 6e– + 8H+ → NH4+ + 2H2O | NIR | Cre09.g410950 | NO3– | [33] | |
| 3 | NH4+ + 2e– + α-ketoglutarate + ATP→ Glutamate + ADP + Pi | GS/GOGAT | [33] | |||
| 4 | NO2– + e– + 2H+ → NO + H2O | NR/NOFNIR NirK | Cre09.g395950/Cre09.g389089 Cre08.g360550 | NO3– Dark | Cytosol Mitochondria | [35] [36] |
| 5 | NO + 2H2O → NO3– + 3e– + 4H+ | NR/THB1 | Cre09.g410950/Cre14.g615400 | NO3– | Cytosol | [37] |
| 6 | 2NO + 2e– + 2H+ → N2O + H2O | CYP55 FLVA FLVB HCP1 HCP2 HCP3 HCP4 | Cre01.g007950 Cre12.g531900 Cre16.g691800 Cre09.g391650 Cre09.g393543 Cre09.g393506 Cre09.g391450 | NO3–/Dark - - NO3–/Dark NO3– NO3–/Dark NO3–/Dark/anaerobiosis | Mitochondria* Chloroplast Chloroplast Chloroplast Mitochondria* Chloroplast Chloroplast | [38] [27] [27] [39] [38] [39] [40] |
| 7 | L-Amino acid +H2O+ O2 → NH4+ + α-keto acid + H2O2 | LAO1 | Cre12.g551353 | Nitrogen starvation | Periplasm | [35] |
| Microalgae | NIRK | HCP | CYP55 | FLVA | FLVB | N2O/Ref. |
|---|---|---|---|---|---|---|
| Chlamydomonas reinhardtii | PNW79625.1 | XP_001694756.1 XP_001694571.1 XP_001694671.1 XP_001694454.1 | XP_001700272.1 | XP_001692916.1 | PNW71243.1 | + [26,27] |
| Chlamydomonas eustigma | GAX74118.1 | GAX73888.1 | GAX73438.1 | |||
| Chlorella variabilis | XP_005851548.1 | XP_005848845.1 | XP 005849742.1 | XP 005849474.1 | + [27] | |
| Chlorella sorokiniana | PRW57688.1 | PRW58172.1 | PRW58571.1 (A) | PRW59370.1 | PRW58882.1 | |
| PRW60486.1 | PRW58572.1 (B) | |||||
| PRW58278.1 | ||||||
| Scenedesmus obliquus UTEX 393 | SZX67391.1 | ID:15469 | ID:435 | SZX62872.1 | SZX63966.1 | + [89] |
| ID:3042 | SZX76545.1 | SZX639665.1 | ||||
| Scenedesmus obliquus EN0004 | ID:579459 | ID:571596 | ID:324854 | ID:321778/332768 | XP002948938.1 | + [89] |
| ID:618262 | ID:342393 | |||||
| Nannochloropsis salina | NSK 004142 | + [90,91] | ||||
| Nannochloropsis oceanica CCMP1779 | 599317 | |||||
| Nannochloropsis gaditana | XP 005853562.1 | − [27] | ||||
| Gonium pectorale | KXZ41847.1 | KXZ42279.1 | KXZ51271.1 | KXZ42718.1 | ||
| KXZ44628.1 | ||||||
| Coccomyxa subelipsoidea | XP 005643451.1 | XP 005643449.1 | + [27] | |||
| Tetrabaena socialis | PNH09786.1 | |||||
| Raphidocelis subcapitata | GBF99305.1 | GBF95475.1 | GBF88523.1 | GBF98453.1 | ||
| GBF96263.1 | ||||||
| Volvox carteri | XP_002952683.1 | XP 002947563.1 | XP 002948938.1 | |||
| Yamagishiella unicocca | ARO50069.1 | |||||
| Micractinium conductrix | PSC73563.1 | PSC76351.1 | PSC76929.1 | |||
| PSC72373.1 | ||||||
| PSC68490.1 | ||||||
| Micromonas pusilla | XP 003057013.1 | XP 003057495.1 | ||||
| Micromonas commoda | XP 002502419.1 | XP 002502038.1 | ||||
| Monoraphidium neglectum SAG 48.87 | XP013900867.1 | ID:2893 ID:2894 | ||||
| Bathycoccus prasinos | XP007514084.1 | XP 007514099.1 | ||||
| Bathycoccus lucimarinus | XP001416100.1 | XP001416098.1 | ||||
| Ostreococcus tauri | XP003075211.2 | XP003075215.2 | ||||
| Klebsormidium nitens | GAQ79906.1 | GAQ89021.1 | ||||
| Galdieria sulphuraria | XP_005707361.1 | − [27] | ||||
| Porphyridium purpureum | − [27] | |||||
| Phaeodactylum tricornutum | ID:12416 | − [27] | ||||
| Thalassiosira pseudonana | ID:32716 | ID:263399a | − [27] | |||
| Guillardia theta CCMP2712 | ID:152637 | |||||
| Andalucia godoyi | KAF0852161.1 | |||||
| Phaeocystis antarctica CCMP1374 | ID:684 | |||||
| Emiliana huxleyi | XP 005783766.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellido-Pedraza, C.M.; Calatrava, V.; Sanz-Luque, E.; Tejada-Jiménez, M.; Llamas, Á.; Plouviez, M.; Guieysse, B.; Fernández, E.; Galván, A. Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle. Plants 2020, 9, 903. https://doi.org/10.3390/plants9070903
Bellido-Pedraza CM, Calatrava V, Sanz-Luque E, Tejada-Jiménez M, Llamas Á, Plouviez M, Guieysse B, Fernández E, Galván A. Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle. Plants. 2020; 9(7):903. https://doi.org/10.3390/plants9070903
Chicago/Turabian StyleBellido-Pedraza, Carmen M., Victoria Calatrava, Emanuel Sanz-Luque, Manuel Tejada-Jiménez, Ángel Llamas, Maxence Plouviez, Benoit Guieysse, Emilio Fernández, and Aurora Galván. 2020. "Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle" Plants 9, no. 7: 903. https://doi.org/10.3390/plants9070903