Mutations of OsPLDa1 Increase Lysophospholipid Content and Enhance Cooking and Eating Quality in Rice
Abstract
:1. Introduction
2. Results
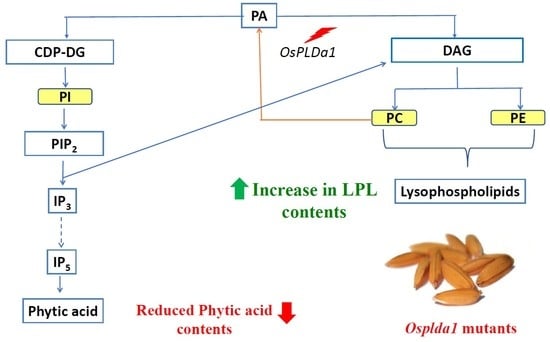
2.1. Lysophospholipid and Gene Expression Analysis of Ospldα1 Mutants
2.2. Apparent Amylose Contents (AAC)
2.3. Thermal and Retrogradation Properties
2.4. Concentration of Phytic Acid in Mutants
2.5. Pasting Properties
3. Discussion
3.1. Association of Lysophospholipid with Phytic acid in Osplda1 Brown Rice Flour
3.2. Ospldα1 Mutations Significantly Affected the Cooking and Eating Properties of Brown Rice
4. Materials and Methods
4.1. Sample Materials
4.2. Metabolite Profiling
4.3. Apparent Amylose Content (AAC)
4.4. RVA Analyses
4.5. Thermal and Retrogradation Properties
4.6. qRT-PCR Assay
4.7. Evaluation of Phytic Acid Content
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Waters, D.L.; Rose, T.J.; Bao, J.; King, G.J. Phospholipids in rice: Significance in grain quality and health benefits: A review. Food Chem. 2013, 139, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Lei, L.; Waters, D.L.; Jin-Song, B.J. Association mapping and marker development of genes for starch lysophospholipid synthesis in rice. Rice Sci. 2016, 23, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Takáč, T.; Novák, D.; Šamaj, J. Recent Advances in the Cellular and Developmental Biology of Phospholipases in Plants. Front. Plant Sci. 2019, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.P.P. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 2005, 139, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Ok, S.H.; Bahn, S.C.; Jang, J.; Oh, S.A.; Park, S.K.; Twell, D.; Ryu, S.B.; Shin, J.S. Endoplasmic reticulum—and golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell 2011, 23, 94–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Yu, X.; Zhang, X.; Yang, L.; Huang, X.; Zhang, J.; Pritchard, H.W.; Li, W. Phospholipase Dα1-mediated phosphatidic acid change is a key determinant of desiccation-induced viability loss in seeds. Plant Cell Environ. 2018, 41, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zien, C.A.; Afitlhile, M.; Welti, R.; Hildebrand, D.F.; Wang, X. Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 2000, 12, 2237–2246. [Google Scholar] [CrossRef] [Green Version]
- Vadovic, P.; Samajova, O.; Takac, T.; Novák, D.; Zapletalova, V.; Colcombet, J.; Samaj, J. Biochemical and genetic interactions of phospholipase D alpha 1 and mitogen-activated protein kinase 3 affect Arabidopsis stress response. Front. Plant Sci. 2019, 10, 275. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Bahn, S.-C.; Wang, G.; Zhang, Y.; Chen, B.; Zhang, Y.; Wang, X.; Zhao, J. PLDα1-knockdown soybean seeds display higher unsaturated glycerolipid contents and seed vigor in high temperature and humidity environments. Biotechnol. Biofuels 2019, 12, 9. [Google Scholar] [CrossRef]
- Devaiah, S.P.; Pan, X.; Hong, Y.; Roth, M.; Welti, R.; Wang, X. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J. 2007, 50, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Galland, M.; He, D.; Lounifi, I.; Arc, E.; Clément, G.; Balzergue, S.; Huguet, S.; Cueff, G.; Godin, B.; Collet, B. An integrated “multi-omics” comparison of embryo and endosperm tissue-specific features and their impact on rice seed quality. Front. Plant Sci. 2017, 8, 1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Arrigo, P.; Servi, S. Synthesis of lysophospholipids. Molecules 2010, 15, 1354–1377. [Google Scholar]
- Osman, C.; Voelker, D.R.; Langer, T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011, 192, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, G.J.; Liu, Q.; Shreshtha, P.; Li, Z.; Rahman, S. RNAi-mediated down-regulation of the expression of OsFAD2-1: Effect on lipid accumulation and expression of lipid biosynthetic genes in the rice grain. BMC Plant Biol. 2016, 16, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J. Rice starch. In Bao Rice Chemistry and Technology, 4th ed.; Elsevier: Amsterdam, The Nederland, 2019; Volume 2, pp. 55–108. [Google Scholar]
- Khan, M.S.S.; Basnet, R.; Islam, S.A.; Shu, Q. Mutational analysis of OsPLDα1 reveals its involvement in phytic acid biosynthesis in rice grains. J. Agric. Food Chem. 2019, 67, 11436–11443. [Google Scholar] [CrossRef]
- Tong, C.; Chen, Y.; Tan, Y.; Liu, L.; Waters, D.L.; Rose, T.J.; Shu, Q.; Bao, J. Analysis of lysophospholipid content in low phytate rice mutants. J. Agric. Food Chem. 2017, 65, 5435–5441. [Google Scholar] [CrossRef]
- Dalmau, N.; Bedia, C.; Tauler, R. Validation of the regions of interest multivariate curve resolution (ROIMCR) procedure for untargeted LC-MS lipidomic analysis. Anal. Chim. 2018, 1025, 80–91. [Google Scholar] [CrossRef]
- Wu, K.; Gunaratne, A.; Gan, R.; Bao, J.; Corke, H.; Jiang, F. Relationships Between Cooking Properties and Physicochemical Properties in Brown and White Rice. Starch 2018, 70, 1700167. [Google Scholar] [CrossRef]
- Ali, W.H.; Chen, Q.; Delgiorno, K.E.; Su, W.; Hall, J.C.; Hongu, T.; Tian, H.; Kanaho, Y.; Di Paolo, G.; Crawford, H.C. Deficiencies of the lipid-signaling enzymes phospholipase D1 and D2 alter cytoskeletal organization, macrophage phagocytosis, and cytokine-stimulated neutrophil recruitment. PLoS ONE 2013, 8, e55325. [Google Scholar] [CrossRef] [Green Version]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P. Acyl-lipid metabolism. TAB 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, G.F.; Paul, R.U.; Holk, A.; Martinec, J. Down-regulation by elicitors of phosphatidylcholine-hydrolyzing phospholipase C and up-regulation of phospholipase A in plant cells. Biochem. Biophys. Res. 2002, 293, 766–770. [Google Scholar] [CrossRef]
- Ryu, S.B.; Lee, H.Y.; Doelling, J.H.; Palta, J.P. Characterization of a cDNA encoding Arabidopsis secretory phospholipase A2-α, an enzyme that generates bioactive lysophospholipids and free fatty acids. Biochim. Biophys. Acta. 2005, 2, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.-J.; Tian, Z.-X.; Fang, Y.-W.; Yang, Y.-C.; Li, J.; Zeng, S.-Y.; Gu, S.-L.; Xu, C.-W.; Tang, S.-Z.; Gu, M.-H. Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 122, 63–76. [Google Scholar] [CrossRef]
- Liu, L.; Tong, C.; Bao, J.; Waters, D.L.E.; Rose, T.J.; King, G.J. Determination of Starch Lysophospholipids in Rice Using Liquid Chromatography–Mass Spectrometry (LC-MS). J. Agric. Food Chem. 2014, 62, 6600–6607. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Liu, L.; Waters, D.L.; Huang, Y.; Bao, J. The contribution of lysophospholipids to pasting and thermal properties of nonwaxy rice starch. Carbohydrol 2015, 133, 187–193. [Google Scholar] [CrossRef]
- Tong, C.; Bao, J. Rice lipids and rice bran oil. Rice 2019, 4, 131–168. [Google Scholar]
- Shu, Q.Y.; Wu, D.X.; Xia, Y.W.; Gao, M.W.; McClung, A. Relationship between RVA profile character and eating quality in Oryza sativa L. Sci. Agric. Sinica 1998, 31, 25–29. [Google Scholar]
- Bao, J.; Corke, H.; He, P.; Zhu, L.H. Analysis of quantitative trait loci for starch properties of rice based on an RIL population. Acta Bot. Sinica 2003, 8, 986–994. [Google Scholar]
- Ahmed, S.; Zhou, X.; Pang, Y.; Xu, Y.; Tong, C.; Bao, J. Genetic diversity of potato genotypes estimated by starch physicochemical properties and microsatellite markers. Food Chem. 2018, 257, 368–375. [Google Scholar] [CrossRef]
- McKie, V.A.; McCleary, B.V. A novel and rapid colorimetric method for measuring total phosphorus and phytic acid in foods and animal feeds. J. AOAC Int. 2016, 99, 738–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Locations | AAC (%) | To (°C) | Tp (°C) | Tc (°C) | ΔHg (j/g) | ΔHr (j/g) | R% | |
|---|---|---|---|---|---|---|---|---|
| Xidao#1 | Hangzhou | 17.17 ± 0.13 a | 62.03 ± 0.27 a | 71.38 ± 0.47 a | 77.95 ± 0.55 a | 5.75 ± 0.41 a | 0.33 ± 0.09 a | 5.91 ± 1.13 a |
| osplda1-1 | 15.33 ± 0.42 b | 60.71 ± 0.34 b | 70.96 ± 0.15 b | 77.22 ± 0.11 a | 7.98 ± 0.87 b | 0.16 ± 0.01 b | 2.11 ± 0.09 b | |
| osplda1-2 | 15.24 ± 0.15b | 60.91 ± 0.33 b | 70.41 ± 0.20 b | 77.45 ± 0.65 a | 7.65 ± 0.51 b | 0.17 ± 0.01 b | 2.3 ± 0.11 b | |
| Xidao#1 | Lingshui | 17.28 ± 0.11a | 61.81 ± 0.21 a | 71.07 ± 0.09 a | 77.90 ± 0.48 a | 5.64 ± 0.28 a | 0.36 ± 0.01 a | 6.51 ± 0.38 a |
| osplda1-1 | 15.39 ± 0.39b | 60.60 ± 0.17 b | 70.33 ± 0.40 b | 77.40 ± 0.31 a | 7.61 ± 0.48 b | 0.17 ± 0.01 b | 2.2 ± 0.17 b | |
| osplda1-2 | 15.4 ± 0.18b | 60.68 ± 0.13 b | 70.47 ± 0.38 b | 77.21 ± 0.29 a | 7.72 ± 0.48 b | 0.17 ± 0.03 b | 2.3 ± 0.16 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.S.S.; Basnet, R.; Ahmed, S.; Bao, J.; Shu, Q. Mutations of OsPLDa1 Increase Lysophospholipid Content and Enhance Cooking and Eating Quality in Rice. Plants 2020, 9, 390. https://doi.org/10.3390/plants9030390
Khan MSS, Basnet R, Ahmed S, Bao J, Shu Q. Mutations of OsPLDa1 Increase Lysophospholipid Content and Enhance Cooking and Eating Quality in Rice. Plants. 2020; 9(3):390. https://doi.org/10.3390/plants9030390
Chicago/Turabian StyleKhan, Muhammad Saad Shoaib, Rasbin Basnet, Sulaiman Ahmed, Jinsong Bao, and Qingyao Shu. 2020. "Mutations of OsPLDa1 Increase Lysophospholipid Content and Enhance Cooking and Eating Quality in Rice" Plants 9, no. 3: 390. https://doi.org/10.3390/plants9030390