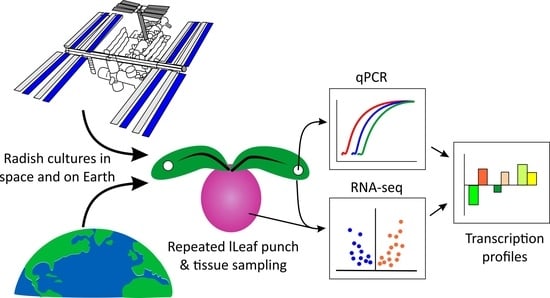
Assessing Radish Health during Space Cultivation by Gene Transcription
Abstract
:1. Introduction
2. Results
2.1. Evaluation of qPCR of LP Samples over Time
2.2. Comparison between SPGE and RNA-Seq Data of Bulbs
2.3. Comparison between Leaf Punch and RNA-Seq Data
2.4. Plant Productivity
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zabel, P.; Bamsey, M.; Schubert, D.; Tajmar, M. Review and analysis of over 40 years of space plant growth systems. Life Sci. Space Res. 2016, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.M. Agriculture for space: People and places paving the way. Open Agric. 2017, 2, 14–32. [Google Scholar] [CrossRef]
- Musgrave, M.E.; Kuang, A.; Xiao, Y.; Stout, S.C.; Bingham, G.E.; Briarty, L.G.; Levinskikh, M.A.; Sychev, V.N.; Podolski, I.G. Gravity independence of seed-to-seed cycling in Brassica rapa. Planta 2000, 210, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Ramonell, K.M.; Mcclure, G.; Musgrave, M.E. Oxygen control of ethylene biosynthesis during seed development in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 2002, 25, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Hasenstein, K.H. Oxygen dependency of germinating Brassica seeds. Life Sci. Space Res. 2016, 8, 30–37. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Abou-Issa, F.; Hasenstein, K.H. Space Flight Cultivation for Radish (Raphanus sativus) in the Advanced Plant Habitat. Gravitational Space Res. 2021, 9, 121–132. [Google Scholar] [CrossRef]
- Monje, O.; Richards, J.T.; Carver, J.A.; Dimapilis, D.I.; Levine, H.G.; Dufour, N.F.; Onate, B.G. Hardware Validation of the Advanced Plant Habitat on ISS: Canopy Photosynthesis in Reduced Gravity. Front. Plant Sci. 2020, 11, 673. [Google Scholar] [CrossRef]
- Kordyum, E.; Hasenstein, K.H. Plant biology for space exploration—Building on the past, preparing for the future. Life Sci. Space Res. 2021, 29, 1–7. [Google Scholar] [CrossRef]
- Ferl, R.; Wheeler, R.; Levine, H.G.; Paul, A.L. Plants in Space. Curr. Opin. Plant Biol. 2002, 5, 258–263. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Brown, C.S.; Levine, H.G.; Krikorian, A.D. Growth and photosynthetic responses of wheat plants crown in space. Plant Physiol. 1996, 110, 801–806. [Google Scholar] [CrossRef]
- Monje, O.; Stutte, G.; Chapman, D. Microgravity does not alter plant stand gas exchange of wheat at moderate light levels and saturating CO2 concentration. Planta 2005, 222, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Kitaya, Y.; Kawai, M.; Tsuruyama, J.; Takahashi, H.; Tani, A.; Goto, E.; Saito, T.; Kiyota, M. The effect of gravity on surface temperature and net photosynthetic rate of plant leaves. Adv. Space Res. 2001, 28, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.-L.; Zhou, M.; Callaham, J.B.; Reyes, M.; Stasiak, M.; Riva, A.; Zupanska, A.K.; Dixon, M.A.; Ferl, R.J. Patterns of Arabidopsis gene expression in the face of hypobaric stress. AoB Plants 2017, 9, plx030. [Google Scholar] [CrossRef]
- Wheeler, R.M.; Wehkamp, C.A.; Stasiak, M.A.; Dixon, M.A.; Rygalov, V.Y. Plants survive rapid decompression: Implications for bioregenerative life support. Adv. Space Res. 2011, 47, 1600–1607. [Google Scholar] [CrossRef]
- Wolff, S.A.; Coelho, L.H.; Zabrodina, M.; Brinckmann, E.; Kittang, A.-I. Plant mineral nutrition, gas exchange and photosynthesis in space: A review. Adv. Space Res. 2013, 51, 465–475. [Google Scholar] [CrossRef]
- Prasad, B.; Richter, P.; Vadakedath, N.; Haag, F.W.M.; Strauch, S.M.; Mancinelli, R.; Schwarzwälder, A.; Etcheparre, E.; Gaume, N.; Lebert, M. How the space environment influences organisms: An astrobiological perspective and review. Int. J. Astrobiol. 2021, 20, 159–177. [Google Scholar] [CrossRef]
- Rea, G.; Esposito, D.; Damasso, M.; Serafini, A.; Margonelli, A.; Faraloni, C.; Torzillo, G.; Zanini, A.; Bertalan, I.; Johanningmeier, U.; et al. Ionizing radiation impacts photochemical quantum yield and oxygen evolution activity of Photosystem II in photosynthetic microorganisms. Int. J. Radiat. Biol. 2008, 84, 867–877. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Grinberg, M.A.; Sukhov, V.; Vodeneev, V. Effect of ionizing radiation on physiological and molecular processes in plants. J. Environ. Radioact. 2019, 202, 8–24. [Google Scholar] [CrossRef]
- Fu, Y.; Li, L.; Xie, B.; Dong, C.; Wang, M.; Jia, B.; Shao, L.; Dong, Y.; Deng, S.; Liu, H.; et al. How to establish a bioregenerative life support system for long-term crewed missions to the Moon or Mars. Astrobiology 2016, 16, 925–936. [Google Scholar] [CrossRef]
- Stankovic, B. Into space—A journey of how humans adapt and live in microgravity. In Plants in Space; IntechOpen: London, UK, 2018; pp. 153–170. [Google Scholar] [CrossRef]
- Wheeler, R.M. Plants for human life support in space: From Myers to Mars. Gravitational Space Res. 2010, 23, 25–35. [Google Scholar]
- Park, M.R.; Wang, Y.-H.; Hasenstein, K.H. Profiling Gene Expression in Germinating Brassica Roots. Plant Mol. Biol. Report. 2014, 32, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Scherp, P.; Hasenstein, K.H. Solid phase gene extraction isolates mRNA at high spatial and temporal resolution. BioTechniques 2008, 45, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Nestorova, G.G.; Crews, N.; Schramm, A.K.; Aquilina, R.A.; Parra, M.; Chin, M.; Chinn, T.; Hee, L. Spaceflight validation of one-step Gene Sampling tool for genetic analysis on the International Space Station. Acta Astronaut. 2022, 198, 225–232. [Google Scholar] [CrossRef]
- Nestorova, G.G.; Hasenstein, K.; Nguyen, N.; DeCoster, M.A.; Crews, N.D. Lab-on-a-chip mRNA purification and reverse transcription via a solid-phase gene extraction technique. Lab Chip 2017, 17, 1128–1136. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Costa-Pérez, A.; Núñez-Gómez, V.; Baenas, N.; Di Pede, G.; Achour, M.; Manach, C.; Mena, P.; Del Rio, D.; García-Viguera, C.; Moreno, D.A.; et al. Systematic Review on the Metabolic Interest of Glucosinolates and Their Bioactive Derivatives for Human Health. Nutrients 2023, 15, 1424. [Google Scholar] [CrossRef]
- Fahey, J.W.; Wehage, S.L.; Holtzclaw, W.D.; Kensler, T.W.; Egner, P.A.; Shapiro, T.A.; Talalay, P. Protection of Humans by Plant Glucosinolates: Efficiency of Conversion of Glucosinolates to Isothiocyanates by the Gastrointestinal Microflora. Cancer Prev. Res. 2012, 5, 603–611. [Google Scholar] [CrossRef]
- Mithen, R.F.; Dekker, M.; Verkerk, R.; Rabot, S.; Johnson, I.T. The nutritional significance, biosynthesis and bioavailability of glucosinolates in human foods. J. Sci. Food Agric. 2000, 80, 967–984. [Google Scholar] [CrossRef]
- Zeng, W.J.; Yang, J.; He, Y.; Zhu, Z.J. Bioactive compounds in cruciferous sprouts and microgreens and the effects of sulfur nutrition. J. Sci. Food Agric. 2023. [Google Scholar] [CrossRef]
- Cai, J.; Li, D.; Aharoni, A. The role of long-distance mobile metabolites in the plant stress response and signaling. Plant J. 2023, 114, 1115–1131. [Google Scholar] [CrossRef]
- Benstein, R.M.; Ludewig, K.; Wulfert, S.; Wittek, S.; Gigolashvili, T.; Frerigmann, H.; Gierth, M.; Flügge, U.-I.; Krueger, S. Arabidopsis Phosphoglycerate Dehydrogenase1 of the Phosphoserine Pathway Is Essential for Development and Required for Ammonium Assimilation and Tryptophan Biosynthesis. Plant Cell 2013, 25, 5011–5029. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Shen, W.; Qian, H.; Zhang, M.; Liu, L.; Wang, Q. Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 5707–5719. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Uddin, M.R.; Park, S.U. Glucosinolate accumulation in three important radish (Raphanus sativus) cultivars. Aust. J. Crop Sci. 2013, 7, 1843–1847. [Google Scholar]
- Rozpądek, P.; Ślesak, I.; Cebula, S.; Waligórski, P.; Dziurka, M.; Skoczowski, A.; Miszalski, Z. Ozone fumigation results in accelerated growth and persistent changes in the antioxidant system of Brassica oleracea L. var. capitata f. alba. J. Plant Physiol. 2013, 170, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Bagchi, R.; Jeschke, V.; Rasmussen, A.R.M.; Hopper, A.; Burow, M.; Estelle, M.; Kliebenstein, D.J. Diverse Allyl Glucosinolate Catabolites Independently Influence Root Growth and Development. Plant Physiol. 2020, 183, 1376–1390. [Google Scholar] [CrossRef]
- Zang, Y.-X.; Kim, H.U.; Kim, J.A.; Lim, M.-H.; Jin, M.; Lee, S.C.; Kwon, S.-J.; Lee, S.-I.; Hong, J.K.; Park, T.-H.; et al. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2009, 276, 3559–3574. [Google Scholar] [CrossRef]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef]
- Pan, Y.; Xu, Y.-y.; Zhu, X.-w.; Liu, Z.; Gong, Y.-q.; Xu, L.; Gong, M.-y.; Liu, L.-w. Molecular Characterization and Expression Profiles of Myrosinase Gene (RsMyr2) in Radish (Raphanus sativus L.). J. Integr. Agric. 2014, 13, 1877–1888. [Google Scholar] [CrossRef]
- Bektas, I.; Fellenberg, C.; Paulsen, H. Water-soluble chlorophyll protein (WSCP) of Arabidopsis is expressed in the gynoecium and developing silique. Planta 2012, 236, 251–259. [Google Scholar] [CrossRef]
- Takahashi, S.; Ono, M.; Uchida, A.; Nakayama, K.; Satoh, H. Molecular cloning and functional expression of a water-soluble chlorophyll-binding protein from Japanese wild radish. J. Plant Physiol. 2013, 170, 406–412. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef]
- Meneguetti, B.T.; Machado, L.D.S.; Oshiro, K.G.N.; Nogueira, M.L.; Carvalho, C.M.E.; Franco, O.L. Antimicrobial peptides from fruits and their potential use as biotechnological tools-A review and outlook. Front. Microbiol. 2017, 7, 2136. [Google Scholar] [CrossRef]
- Muren, E.; Ek, B.; Bjork, I.; Rask, L. Structural comparison of the precursor and the mature form of napin, the 2S storage protein in Brassica napus. Eur. J. Biochem. 1996, 242, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Kim, W.-S.; Song, B.; Oehrle, N.W.; Liu, S.; Krishnan, H.B. Soybean Mutants Lacking Abundant Seed Storage Proteins Are Impaired in Mobilization of Storage Reserves and Germination. ACS Omega 2020, 5, 8065–8075. [Google Scholar] [CrossRef] [PubMed]
- Decadal Survey. Recapturing a Future for Space Exploration: Life and Physical Sciences Research for a New Era; The National Academies Press: Washington, DC, USA, 2011.
- Ajala, C.; Hasenstein, K.H. Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa. Plants 2022, 11, 1191. [Google Scholar] [CrossRef]
- Aguiar, V.R.C.; Castelli, E.C.; Single, R.M.; Bashirova, A.; Ramsuran, V.; Kulkarni, S.; Augusto, D.G.; Martin, M.P.; Gutierrez-Arcelus, M.; Carrington, M.; et al. Comparison between qPCR and RNA-seq reveals challenges of quantifying HLA expression. Immunogenetics 2023, 75, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Choi, W.-G.; Barker, R.J.; Kim, S.-H.; Swanson, S.J.; Gilroy, S. Variation in the transcriptome of different ecotypes of Arabidopsis thaliana reveals signatures of oxidative stress in plant responses to spaceflight. Am. J. Bot. 2019, 106, 123–136. [Google Scholar] [CrossRef]
- Paul, A.L.; Ferl, R.J. Molecular aspects of stress-gene regulation during spaceflight. J. Plant Growth Regul. 2002, 21, 166–176. [Google Scholar] [CrossRef]
- Khodadad, C.L.; Hummerick, M.E.; Spencer, L.E.; Dixit, A.R.; Richards, J.T.; Romeyn, M.W.; Smith, T.M.; Wheeler, R.M.; Massa, G.D. Microbiological and nutritional analysis of lettuce crops grown on the International Space Station. Front. Plant Sci. 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chhapekar, S.S.; Rameneni, J.J.; Kim, S.; Gan, T.H.; Choi, S.R.; Lim, Y.P. Identification of QTLs and Candidate Genes Related to Flower Traits and Bolting Time in Radish (Raphanus sativus L.). Agronomy 2021, 11, 1623. [Google Scholar] [CrossRef]
- Jung, W.Y.; Park, H.J.; Lee, A.; Lee, S.S.; Kim, Y.-S.; Cho, H.S. Identification of Flowering-Related Genes Responsible for Differences in Bolting Time between Two Radish Inbred Lines. Front. Plant Sci. 2016, 7, 1844. [Google Scholar] [CrossRef] [PubMed]
- Pombo, M.A.; Zheng, Y.; Fei, Z.; Martin, G.B.; Rosli, H.G. Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem. Sci. Rep. 2017, 7, 44905. [Google Scholar] [CrossRef]
- Rachinger, N.; Fischer, S.; Böhme, I.; Linck-Paulus, L.; Kuphal, S.; Kappelmann-Fenzl, M.; Bosserhoff, A.K. Loss of Gene Information: Discrepancies between RNA Sequencing, cDNA Microarray, and qRT-PCR. Int. J. Mol. Sci. 2021, 22, 9349. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasenstein, K.H.; John, S.P.; Vandenbrink, J.P. Assessing Radish Health during Space Cultivation by Gene Transcription. Plants 2023, 12, 3458. https://doi.org/10.3390/plants12193458
Hasenstein KH, John SP, Vandenbrink JP. Assessing Radish Health during Space Cultivation by Gene Transcription. Plants. 2023; 12(19):3458. https://doi.org/10.3390/plants12193458
Chicago/Turabian StyleHasenstein, Karl H., Susan P. John, and Joshua P. Vandenbrink. 2023. "Assessing Radish Health during Space Cultivation by Gene Transcription" Plants 12, no. 19: 3458. https://doi.org/10.3390/plants12193458