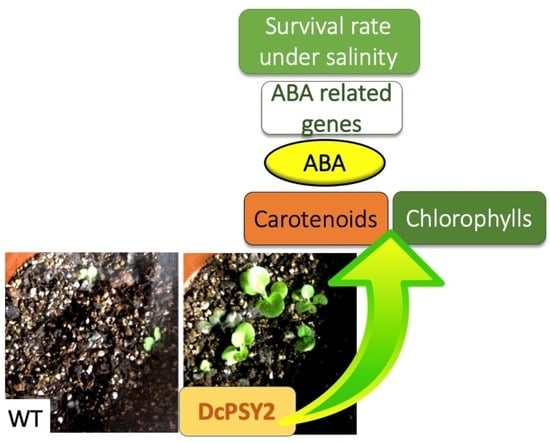
The Carrot Phytoene Synthase 2 (DcPSY2) Promotes Salt Stress Tolerance through a Positive Regulation of Abscisic Acid and Abiotic-Related Genes in Nicotiana tabacum
Abstract
:1. Introduction
2. Methodology
2.1. PSY2 In Silico Analysis
2.2. Vector Construction
2.3. Plant-Material- and Agrobacterium-Mediated Transformation
2.4. RNA Extraction and Quantitative RT-PCR
2.5. Carotenoid and Chlorophyll Extraction and High-Performance Liquid Chromatography (HPLC-DAD)
2.6. Norflurazon Treatment
2.7. Salt Stress Tolerance Assay
2.8. ABA Quantification
3. Results
3.1. DcPSY2 In Silico Analysis and Expression during Carrot Development
3.2. Expression of DcPSY2 in N. tabacum Promotes an Increment in Carotenoid Content
3.3. Transgenic Tobacco DcPSY2 Lines Showed Salt Stress Tolerance in a Chronic Salt Stress Assay
3.4. DcPSY2 Transgenic Plants Induce a Boost in ABA and Stress-Associated Genes under Acute Salt Treatment
3.5. Endogenous NtPSY Gene Expression Is Enhanced in DcPSY2 Transgenic Lines after Salt Stress Treatment
4. Discussion
4.1. DcPSY2 Boost Carotenoid, Phytoene and Chlorophyll Content in N. tabacum
4.2. Constitutive Expression of DcPSY2 Leads to an Increase in Salt Tolerance and ABA Levels in Transgenic Tobacco
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Stange, C.; Flores, C. Carotenoids and Photosynthesis—Regulation of Carotenoid Biosyntesis by Photoreceptors. In Advances Photosynthesis-Fundamental Aspects; Mahadi, M., Ed.; INTECH: Rijeka, Croatia, 2012; pp. 77–96. [Google Scholar]
- Howitt, C.A.; Pogson, B.J. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell. Environ. 2006, 29, 435–445. [Google Scholar] [CrossRef]
- Woodall, A.A.; Lee, S.W.; Weesie, R.J.; Jackson, M.J.; Britton, G. Oxidation of carotenoids by free radicals: Relationship between structure and reactivity. Biochim. Biophys. Acta 1997, 1336, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Demmig-Adams, B.; Gilmore, A.M.; Adams, W.W., 3rd. Carotenoids 3: In vivo function of carotenoids in higher plants. FASEB J. 1996, 10, 403–412. [Google Scholar] [CrossRef]
- Nelson, J.L.; Bernstein, P.S.; Schmidt, M.C.; Von Tress, M.S.; Askew, E.W. Dietary modification and moderate antioxidant supplementation differentially affect serum carotenoids, antioxidant levels and markers of oxidative stress in older humans. J. Nutr. 2003, 133, 3117–3123. [Google Scholar] [CrossRef] [PubMed]
- Vachali, P.; Li, B.; Nelson, K.; Bernstein, P.S. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch. Biochem. Biophys. 2012, 519, 32–37. [Google Scholar] [CrossRef]
- Xie, X.; Yoneyama, K.; Yoneyama, K. The strigolactone story. Ann. Rev. Phytopathol. 2010, 48, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G.; Al-Babili, S.; von Lintig, J. Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 2003, 8, 145–149. [Google Scholar] [CrossRef]
- Lois, L.M.; Rodriguez-Concepcion, M.; Gallego, F.; Campos, N.; Boronat, A. Carotenoid biosynthesis during tomato fruit development: Regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J. 2000, 22, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Wurtzel, E.T. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008, 146, 1333–1345. [Google Scholar] [CrossRef]
- Rodriguez-Villalon, A.; Gas, E.; Rodriguez-Concepcion, M. Colors in the dark: A model for the regulation of carotenoid biosynthesis in etioplasts. Plant Signal. Behav. 2009, 4, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Von Lintig, J.; Welsch, R.; Bonk, M.; Giuliano, G.; Batschauer, A.; Kleinig, H. Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J. 1997, 12, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Bartley, G.E.; Scolnik, P.A. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 1993, 268, 25718–25721. [Google Scholar] [CrossRef]
- Karvouni, Z.; John, I.; Taylor, J.E.; Watson, C.F.; Turner, A.J.; Grierson, D. Isolation and characterisation of a melon cDNA clone encoding phytoene synthase. Plant Mol. Biol. 1995, 27, 1153–1162. [Google Scholar] [CrossRef]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Zhu, Y.; Graham, J.E.; Ludwig, M.; Xiong, W.; Alvey, R.M.; Shen, G.; Bryant, D.A. Roles of xanthophyll carotenoids in protection against photoinhibition and oxidative stress in the cyanobacterium Synechococcus sp. strain PCC 7002. Arch. Biochem. Biophys. 2010, 504, 86–99. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Rodriguez-Concepcion, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Am. Soc. Plant Biol. 2012, 10, e0158. [Google Scholar] [CrossRef]
- Rosas-Saavedra, C.; Stange, C. Biosynthesis of Carotenoids in Plants: Enzymes and Color. Sub-Cell. Biochem. 2016, 79, 35–69. [Google Scholar] [CrossRef]
- Zhu, C.; Bai, C.; Sanahuja, G.; Yuan, D.; Farre, G.; Naqvi, S.; Shi, L.; Capell, T.; Christou, P. The regulation of carotenoid pigmentation in flowers. Arch. Biochem. Biophys. 2010, 504, 132–141. [Google Scholar] [CrossRef]
- Bartley, G.E.; Viitanen, P.V.; Bacot, K.O.; Scolnik, P.A. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 1992, 267, 5036–5039. [Google Scholar] [CrossRef]
- Scolnik, P.A.; Bartley, G.E. Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol. 1994, 104, 1471–1472. [Google Scholar] [CrossRef] [PubMed]
- Buckner, B.; Miguel, P.S.; Janick-Buckner, D.; Bennetzen, J.L. The y1 gene of maize codes for phytoene synthase. Genetics 1996, 143, 479–488. [Google Scholar] [CrossRef]
- Qin, X.; Coku, A.; Inoue, K.; Tian, L. Expression, subcellular localization, and cis-regulatory structure of duplicated phytoene synthase genes in melon (Cucumis melo L.). Planta 2011, 234, 737–748. [Google Scholar] [CrossRef]
- Schledz, M.; al-Babili, S.; von Lintig, J.; Haubruck, H.; Rabbani, S.; Kleinig, H.; Beyer, P. Phytoene synthase from Narcissus pseudonarcissus: Functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J. 1996, 10, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Romer, S.; Hugueney, P.; Bouvier, F.; Camara, B.; Kuntz, M. Expression of the genes encoding the early carotenoid biosynthetic enzymes in Capsicum annuum. Biochem. Biophys. Res. Commun. 1993, 196, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Arango, J.; Wust, F.; Beyer, P.; Welsch, R. Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 2010, 232, 1251–1262. [Google Scholar] [CrossRef]
- Busch, M.; Seuter, A.; Hain, R. Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol. 2002, 128, 439–453. [Google Scholar] [CrossRef]
- Welsch, R.; Wust, F.; Bar, C.; Al-Babili, S.; Beyer, P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008, 147, 367–380. [Google Scholar] [CrossRef]
- Fraser, P.D.; Kiano, J.W.; Truesdale, M.R.; Schuch, W.; Bramley, P.M. Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol. Biol. 1999, 40, 687–698. [Google Scholar] [CrossRef]
- Giorio, G.; Stigliani, A.L.; D’Ambrosio, C. Phytoene synthase genes in tomato (Solanumlycopersicum L.)—new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008, 275, 527–535. [Google Scholar] [CrossRef]
- Li, F.; Tsfadia, O.; Wurtzel, E.T. The phytoene synthase gene family in the Grasses: Subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal. Behav. 2009, 4, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Tzfadia, O.; Vallabhaneni, R.; Gehring, C.; Wurtzel, E.T. A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst. Biol. 2011, 5, 77. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Arbona, V.; Gomez-Cadenas, A.; Rodriguez-Concepcion, M.; Rodriguez-Villalon, A. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in arabidopsis. PLoS ONE 2014, 9, e90765. [Google Scholar] [CrossRef] [PubMed]
- Just, B.J.; Santos, C.A.; Fonseca, M.E.; Boiteux, L.S.; Oloizia, B.B.; Simon, P.W. Carotenoid biosynthesis structural genes in carrot (Daucus carota): Isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theor. Appl. Genet. 2007, 114, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Just, B.J.; Santos, C.A.; Yandell, B.S.; Simon, P.W. Major QTL for carrot color are positionally associated with carotenoid biosynthetic genes and interact epistatically in a domesticated x wild carrot cross. Theor. Appl. Genet. 2009, 119, 1155–1169. [Google Scholar] [CrossRef]
- Fuentes, P.; Pizarro, L.; Moreno, J.C.; Handford, M.; Rodriguez-Concepcion, M.; Stange, C. Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol. Biol. 2012, 79, 47–59. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Stange, C. Biosynthesis of carotenoids in carrot: An underground story comes to light. Arch. Biochem. Biophys. 2013, 539, 110–116. [Google Scholar] [CrossRef]
- Wang, H.; Ou, C.G.; Zhuang, F.Y.; Ma, Z.G. The dual role of phytoene synthase genes in carotenogenesis in carrot roots and leaves. Mol. Breed 2014, 34, 2065–2079. [Google Scholar] [CrossRef]
- Simpson, K.; Fuentes, P.; Quiroz-Iturra, L.F.; Flores-Ortiz, C.; Contreras, R.; Handford, M.; Stange, C. Unraveling the induction of phytoene synthase 2 expression by salt stress and abscisic acid in Daucus carota. J. Exp. Bot. 2018, 69, 4113–4126. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef]
- Horsch, R.B.; Rogers, S.G.; Fraley, R.T. Transgenic plants. Cold Spring Harbor Symp. Quant. Biol. 1985, 50, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Saha, D.; Mondal, T.K. An optimized method for extraction of RNA from tea roots for functional genomics analysis. Indian J. Biotechnol. 2013, 12, 129–132. [Google Scholar]
- Moreno, J.C.; Pizarro, L.; Fuentes, P.; Handford, M.; Cifuentes, V.; Stange, C. Levels of lycopene beta-cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS ONE 2013, 8, e58144. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Britton, G. Structure and properties of carotenoids in relation to function. Faseb J. 1995, 9, 1551–1558. [Google Scholar] [CrossRef]
- Britton, G.; Liaaen-Jensen, H.; Pfander, H. Carotenoids Handbook; Britton, G., Liaaen-Jensen, H., Pfander, H., Eds.; Birkhauser Verlag: Basel, Switzerland, 2004. [Google Scholar]
- Simkin, A.J.; Laboure, A.M.; Kuntz, M.; Sandmann, G. Comparison of carotenoid content, gene expression and enzyme levels in tomato (Lycopersicon esculentum) leaves. Z. Nat. C 2003, 58, 371–380. [Google Scholar] [CrossRef]
- Gonzalez, A.; Contreras, R.A.; Zuiga, G.; Moenne, A. Oligo-carrageenan kappa-induced reducing redox status and activation of TRR/TRX system increase the level of indole-3-acetic acid, gibberellin A3 and trans-zeatin in Eucalyptus globulus trees. Molecules 2014, 19, 12690–12698. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.; Tham, C.; Duan, L.; Dinneny, J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef]
- Arias, D.; Arenas, M.A.; Flores-Ortiz, C.; Peirano, C.; Handford, M.; Stange, C. Daucus carota DcPSY2 and DcLCYB1 as Tools for Carotenoid Metabolic Engineering to Improve the Nutritional Value of Fruits. Front. Plant Sci. 2021, 12, 677553. [Google Scholar] [CrossRef]
- Hartel, H.; Grimm, B. Consequences of chlorophyll deficiency for leaf carotenoid composition in tobacco synthesizing glutamate 1-semialdehyde aminotransferase antisense RNA: Dependency on developmental age and growth light. J. Exp. Bot. 1998, 49, 535–546. [Google Scholar] [CrossRef]
- Maass, D.; Arango, J.; Wust, F.; Beyer, P.; Welsch, R. Carotenoid Crystal Formation in Arabidopsis and Carrot Roots Caused by Increased Phytoene Synthase Protein Levels. PLoS ONE 2009, 4, 373. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.C.; Cerda, A.; Simpson, K.; Lopez-Diaz, I.; Carrera, E.; Handford, M.; Stange, C. Increment in Nicotiana tabacum Fitness through Positive Regulation of Carotenoid, Gibberellin and Chlorophyll Pathways promoted by Daucus carota Lycopene β-cyclase (Dclcyb1) expression. J. Exp. Bot. 2016, 2016, 37. [Google Scholar]
- Fray, R.G.; Wallace, A.; Fraser, P.D.; Valero, D.; Hedden, P.; Bramley, P.M.; Grierson, D. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 1995, 8, 693–701. [Google Scholar] [CrossRef]
- Teixeira, E.I.; Fischer, G.; van Velthuizen, H.; Walter, C.; Ewert, F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Cidade, L.C.; de Oliveira, T.M.; Mendes, A.F.; Macedo, A.F.; Floh, E.I.; Gesteira, A.S.; Soares-Filho, W.S.; Costa, M.G. Ectopic expression of a fruit phytoene synthase from Citrus paradisi Macf. promotes abiotic stress tolerance in transgenic tobacco. Mol. Biol. Rep. 2012, 39, 10201–10209. [Google Scholar] [CrossRef]
- Han, H.P.; Li, Y.X.; Zhou, S.F. Overexpression of phytoene synthase gene from Salicornia europaea alters response to reactive oxygen species under salt stress in transgenic Arabidopsis. Biotechnol. Lett. 2008, 30, 1501–1507. [Google Scholar] [CrossRef]
- Fricke, W.; Peters, W.S. The biophysics of leaf growth in salt-stressed barley. A study at the cell level. Plant Physiol. 2002, 129, 374–388. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Du, Y.L.; Wang, Z.Y.; Fan, J.W.; Turner, N.C.; He, J.; Wang, T.; Li, F.M. Exogenous abscisic acid reduces water loss and improves antioxidant defence, desiccation tolerance and transpiration efficiency in two spring wheat cultivars subjected to a soil water deficit. Funct. Plant Biol. 2013, 40, 494–506. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acevedo, O.; Contreras, R.A.; Stange, C. The Carrot Phytoene Synthase 2 (DcPSY2) Promotes Salt Stress Tolerance through a Positive Regulation of Abscisic Acid and Abiotic-Related Genes in Nicotiana tabacum. Plants 2023, 12, 1925. https://doi.org/10.3390/plants12101925
Acevedo O, Contreras RA, Stange C. The Carrot Phytoene Synthase 2 (DcPSY2) Promotes Salt Stress Tolerance through a Positive Regulation of Abscisic Acid and Abiotic-Related Genes in Nicotiana tabacum. Plants. 2023; 12(10):1925. https://doi.org/10.3390/plants12101925
Chicago/Turabian StyleAcevedo, Orlando, Rodrigo A. Contreras, and Claudia Stange. 2023. "The Carrot Phytoene Synthase 2 (DcPSY2) Promotes Salt Stress Tolerance through a Positive Regulation of Abscisic Acid and Abiotic-Related Genes in Nicotiana tabacum" Plants 12, no. 10: 1925. https://doi.org/10.3390/plants12101925