Environmental Niche Dynamics of Blue Grama (Bouteloua gracilis) Ecotypes in Northern Mexico: Genetic Structure and Implications for Restoration Management
Abstract
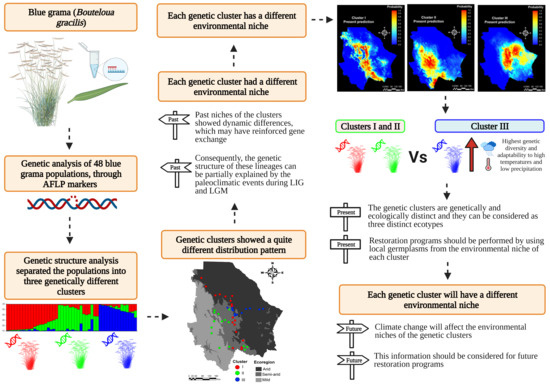
:1. Introduction
2. Results
2.1. Genetic Structure
2.2. Environmental Niche of the Genetic Clusters
3. Discussion
3.1. Genetic Structure
3.2. Environmental Niche Modeling
4. Materials and Methods
4.1. Population Sampling
4.2. Genetic Structure Analysis
4.3. Environmental Niche Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jiménez-Mejías, P.; Fernández-Mazuecos, M.; Amat, M.E.; Vargas, P. Narrow endemics in European mountains: High genetic diversity within the monospecific genus Pseudomisopates (Plantaginaceae) despite isolation since the late Pleistocene. J. Biogeogr. 2015, 42, 1455–1468. [Google Scholar] [CrossRef]
- Sillero, N.; Arenas-Castro, S.; Enriquez-Urzelai, U.; Vale, C.G.; Sousa-Guedes, D.; Martínez-Freiría, F.; Real, R.; Barbosa, A. Want to model a species niche? A step-by-step guideline on correlative ecological niche modelling. Ecol. Model. 2021, 456, 109671. [Google Scholar] [CrossRef]
- Jay, F.; Manel, S.; Alvarez, N.; Durand, E.Y.; Thuiller, W.; Holderegger, R.; Taberlet, P.; François, O. Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 2013, 21, 2354–2368. [Google Scholar] [CrossRef]
- Pauls, S.U.; Nowak, C.; Bálint, M.; Pfenninger, M. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 2012, 22, 925–946. [Google Scholar] [CrossRef]
- Ikeda, D.H.; Max, T.L.; Allan, G.J.; Lau, M.K.; Shuster, S.M.; Whitham, T.G. Genetically informed ecological niche models improve climate change predictions. Glob. Chang. Biol. 2017, 23, 164–176. [Google Scholar] [CrossRef]
- Bothwell, H.M.; Evans, L.M.; Hersch-Green, E.I.; Woolbright, S.A.; Allan, G.J.; Whitham, T.G. Genetic data improves niche model discrimination and alters the direction and magnitude of climate change forecasts. Ecol. Appl. 2021, 31, e2254. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Godsoe, W.; Rodriguez-Sanchez, F.; Wang, H.-H.; Warren, D. Niche Estimation above and below the Species Level. Trends Ecol. Evol. 2019, 34, 260–273. [Google Scholar] [CrossRef]
- Smýkal, P.; Trněný, O.; Brus, J.; Hanáček, P.; Rathore, A.; Roma, R.D.; Pechanec, V.; Duchoslav, M.; Bhattacharyya, D.; Bariotakis, M. Genetic structure of wild pea (Pisum sativum subsp. elatius) populations in the northern part of the Fertile Crescent reflects moderate cross-pollination and strong effect of geographic but not environmental distance. PLoS ONE 2018, 13, e0194056. [Google Scholar]
- Álvarez-Holguín, A.; Morales-Nieto, C.R.; Corrales-Lerma, R.; Prieto-Amparán, J.A.; Villarreal-Guerrero, F.; Sánchez-Gutiérrez, R.A. Genetic structure and temporal environmental niche dynamics of sideoats grama [Bouteloua curtipendula (Michx.) Torr.] populations in Mexico. PLoS ONE 2021, 16, e0254566. [Google Scholar] [CrossRef]
- Hällfors, M.H.; Liao, J.; Dzurisin, J.; Grundel, R.; Hyvärinen, M.; Towle, K.; Wu, G.C.; Hellmann, J.J. Addressing potential local adaptation in species distribution models: Implications for conservation under climate change. Ecol. Appl. 2016, 26, 1154–1169. [Google Scholar] [CrossRef]
- Marcer, A.; Méndez-Vigo, B.; Alonso-Blanco, C.; Picó, F.X. Tackling intraspecific genetic structure in distribution models better reflects species geographical range. Ecol. Evol. 2016, 6, 2084–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banta, J.A.; Ehrenreich, I.M.; Gerard, S.; Chou, L.; Wilczek, A.; Schmitt, J.; Kover, P.X.; Purugganan, M.D. Climate envelope modelling reveals intraspecific relationships among flowering phenology, niche breadth and potential range size in Arabidopsis thaliana. Ecol. Lett. 2012, 15, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Freiría, F.; Freitas, I.; Zuffi, M.A.L.; Golay, P.; Ursenbacher, S.; Velo-Antón, G. Climatic refugia boosted allopatric diversification in Western Mediterranean vipers. J. Biogeogr. 2020, 47, 1698–1713. [Google Scholar] [CrossRef]
- Maguire, K.C.; Shinneman, D.J.; Potter, K.M.; Hipkins, V.D. Intraspecific Niche Models for Ponderosa Pine (Pinus ponderosa) Suggest Potential Variability in Population-Level Response to Climate Change. Syst. Biol. 2018, 67, 965–978. [Google Scholar] [CrossRef]
- Scoble, J.; Lowe, A.J. A case for incorporating phylogeography and landscape genetics into species distribution modelling approaches to improve climate adaptation and conservation planning. Divers. Distrib. 2010, 16, 343–353. [Google Scholar] [CrossRef]
- Mccollum, F.T.; Galyean, M.L.; Krysl, L.J.; Wallace, J.D. Cattle Grazing Blue Grama Rangeland I. Seasonal Diets and Rumen Fermentation. J. Range Manag. 1985, 38, 539–543. [Google Scholar] [CrossRef]
- Morales Nieto, C.R.; Madrid Pérez, L.; Melgoza Castillo, A.; Martínez Salvador, M.; Jurado Guerra, P.; Arévalo Gallegos, S.; Rascón Cruz, Q. Análisis morfológico de la diversidad del pasto navajita [Bouteloua gracilis (Willd. ex Kunth) Lag. ex Steud.], en Chihuahua, México. Téc. Pecu. Méx. 2009, 47, 245–256. [Google Scholar]
- Arredondo Moreno, T.; Huber Sannwald, E.; García Moya, E.; García Holguín, M.; Aguado Santacruz, G.A. Selección de germoplasma de zacate navajita con diferente historial de uso en Jalisco, México. Rev. Mex. Pecu. 2005, 43, 371–385. [Google Scholar]
- Beltrán López, S.; García Díaz, C.A.; Hernández Alatorre, J.A.; Loredo Osti, C.; Urrutia Morales, J.; González Eguiarte, L.A.; Gámez Vázquez, H.G. “Navajita Cecilia” Bouteloua gracilis HBK (Lag.): Nueva variedad de pasto para zonas áridas y semiáridas. Rev. Mex. Pecu. 2010, 1, 127–130. [Google Scholar]
- Morales-Nieto, C.R.; Álvarez-Holguín, A.; Villarreal-Guerrero, F.; Corrales-Lerma, R.; Pinedo-Álvarez, A.; Martínez Salvador, M. Phenotypic and genetic diversity of blue grama (Bouteloua gracilis) populations from Northern Mexico. Arid. Land Res. Manag. 2020, 34, 83–98. [Google Scholar] [CrossRef]
- Vieira, J.; Schnadelbach, A.S.; Hughes, F.M.; Jardim, J.G.; Clark, L.G.; De Oliveira, R.P. Ecological niche modelling and genetic diversity of Anomochloa marantoidea (Poaceae): Filling the gaps for conservation in the earliest-diverging grass subfamily. Bot. J. Linn. Soc. 2020, 192, 258–280. [Google Scholar] [CrossRef]
- Lv, T.; Harris, A.; Liu, Y.; Liu, T.; Liang, R.; Ma, Z.; Su, X. Population genetic structure and evolutionary history of Psammochloa villosa (Trin.) Bor (Poaceae) revealed by AFLP marker. Ecol. Evol. 2021, 11, 10258–10276. [Google Scholar] [CrossRef] [PubMed]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Martinson, E.J.; Eddy, Z.B.; Commerford, J.L.; Blevins, E.; Rolfsmeier, S.J.; Mclauchlan, K.K. Biogeographic Distributions of Selected North American Grassland Plant Species. Phys. Geogr. 2011, 32, 583–602. [Google Scholar] [CrossRef]
- Puga, N.D.; Sifuentes, J.M.; Corral, J.R.; Eguiarte, D.G.; Munguía, S.M. Impacto del cambio climático en las áreas con aptitud ambiental para Bouteloua gracilis y Bouteloua repens en México. Rev. Bio Cienc. 2020, 7, 14. [Google Scholar]
- Schoener, T.W. The Anolis Lizards of Bimini: Resource Partitioning in a Complex Fauna. Ecology 1968, 49, 704–726. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef]
- Tso, K.L.; Allan, G.J. Environmental variation shapes genetic variation in Bouteloua gracilis: Implications for restoration management of natural populations and cultivated varieties in the southwestern United States. Ecol. Evol. 2019, 9, 482–499. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.-H.; Gao, X.-Y.; Huang, J.-F.; Zhang, M.-L. Plant phylogeography in arid Northwest China: Retrospectives and perspectives. J. Syst. Evol. 2015, 53, 33–46. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Fan, Y.; Sun, M.; Wu, W.; Zhao, W.; Yang, X.; Huang, L.; Peng, Y.; Ma, X.; et al. Genetic Structure and Eco-Geographical Differentiation of Wild Sheep Fescue (Festuca ovina L.) in Xinjiang, Northwest China. Molecules 2017, 22, 1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, A.M.; Bushey, J.A.; Ocheltree, T.W.; Smith, M.D. Genetic and functional variation across regional and local scales is associated with climate in a foundational prairie grass. New Phytol. 2020, 227, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Xiong, Y.; Yu, Q.; Zhao, J.; Lei, X.; Dong, Z.; Yang, J.; Song, S.; Peng, Y.; Liu, W.; et al. Genetic variability and structure of an important wild steppe grass Psathyrostachys juncea (Triticeae: Poaceae) germplasm collection from north and central Asia. PeerJ 2020, 8, e9033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.-D.; Liu, W.-H.; Sun, M.; Zhou, J.-Q.; Liu, W.; Zhang, C.-L.; Zhang, X.-Q.; Peng, Y.; Huang, L.-K.; Ma, X. Genetic diversity and structure of Elymus tangutorum accessions from western China as unraveled by AFLP markers. Hereditas 2019, 156, 8. [Google Scholar] [CrossRef]
- Coppi, A.; Lastrucci, L.; Cappelletti, D.; Cerri, M.; Ferranti, F.; Ferri, V.; Foggi, B.; Gigante, D.; Venanzoni, R.; Viciani, D.; et al. AFLP Approach Reveals Variability in Phragmites australis: Implications for Its Die-Back and Evidence for Genotoxic Effects. Front. Plant Sci. 2018, 9, 386. [Google Scholar] [CrossRef]
- Mitchell, M.L.; Stodart, B.J.; Virgona, J.M. Genetic diversity within a population of Microlaena stipoides, as revealed by AFLP markers. Aust. J. Bot. 2015, 62, 580–586. [Google Scholar] [CrossRef]
- Reisch, C.; Anke, A.; Röhl, M. Molecular variation within and between ten populations of Primula farinosa (Primulaceae) along an altitudinal gradient in the northern Alps. Basic Appl. Ecol. 2005, 6, 35–45. [Google Scholar] [CrossRef]
- Kiambi, D.; Newbury, H.; Maxted, N.; Ford-Lloyd, B. Molecular genetic variation in the African wild rice Oryza longistaminata A. Chev. et Roehr. and its association with environmental variables. Afr. J. Biotechnol. 2008, 7, 1446–1460. [Google Scholar]
- Zhao, N.-X.; Gao, Y.-B.; Wang, J.-L.; Ren, A.-Z. Genetic Diversity and Population Differentiation of the Dominant Species Stipa krylovii in the Inner Mongolia Steppe. Biochem. Genet. 2006, 44, 504–517. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, M.; Zhang, X.; Chen, S.; Nie, G.; Peng, Y.; Huang, L.; Ma, X. AFLP-based genetic diversity of wild orchardgrass germplasm collections from Central Asia and Western China, and the relation to environmental factors. PLoS ONE 2018, 13, e0195273. [Google Scholar] [CrossRef] [Green Version]
- Wanjala, B.W.; Obonyo, M.; Wachira, F.N.; Muchugi, A.; Mulaa, M.; Harvey, J.; Skilton, R.A.; Proud, J.; Hanson, J. Genetic diversity in Napier grass (Pennisetum purpureum) cultivars: Implications for breeding and conservation. AoB Plants 2013, 5, plt022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, J.; Wu, Y.; Wang, Z.; Samuels, T. Genetic diversity in tetraploid switchgrass revealed by AFLP marker polymorphisms. Genet. Mol. Res. 2011, 10, 2976–2986. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.J.; Diefendorf, A.F.; Freimuth, E.J.; Hyun, S. Last interglacial (MIS 5e) and Holocene paleohydrology and paleovegetation of midcontinental North America from Gulf of Mexico sediments. Quat. Sci. Rev. 2020, 227, 106066. [Google Scholar] [CrossRef]
- Lüthi, D.; Le Floch, M.; Bereiter, B.; Blunier, T.; Barnola, J.-M.; Siegenthaler, U.; Raynaud, D.; Jouzel, J.; Fischer, H.; Kawamura, K.; et al. High-resolution carbon dioxide concentration record 650,000–800,000 years before present. Nature 2008, 453, 379–382. [Google Scholar] [CrossRef]
- Dutton, A.; Lambeck, K. Ice Volume and Sea Level During the Last Interglacial. Science 2012, 337, 216–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kukla, G.; Bender, M.; De Beaulieu, J.; Bond, G.; Broecker, W.; Cleveringa, P.; Gavin, J.E.; Herbert, T.D.; Imbrie, J.; Jouzel, J.; et al. Last Interglacial Climates. Quat. Res. 2002, 58, 2–13. [Google Scholar] [CrossRef]
- Yin, Q.Z.; Berger, A. Individual contribution of insolation and CO2 to the interglacial climates of the past 800,000 years. Clim. Dyn. 2012, 38, 709–724. [Google Scholar] [CrossRef]
- Gugger, P.F.; Ikegami, M.; Sork, V.L. Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol. Ecol. 2013, 22, 3598–3612. [Google Scholar] [CrossRef]
- Kitoh, A.; Murakami, S. Tropical Pacific climate at the mid-Holocene and the Last Glacial Maximum simulated by a coupled ocean-atmosphere general circulation model. Paleoceanography 2002, 17, 19-1–19-13. [Google Scholar] [CrossRef]
- Otto-Bliesner, B.L.; Marshall, S.J.; Overpeck, J.T.; Miller, G.H.; Hu, A.; CAPE Last Interglacial Project Members. Simulating Arctic Climate Warmth and Icefield Retreat in the Last Interglaciation. Science 2006, 311, 1751–1753. [Google Scholar] [CrossRef] [Green Version]
- Bigelow, N.H.; Brubaker, L.B.; Edwards, M.E.; Harrison, S.; Prentice, I.C.; Anderson, P.M.; Andreev, A.; Bartlein, P.; Christensen, T.; Cramer, W.; et al. Climate change and Arctic ecosystems: 1. Vegetation changes north of 55° N between the last glacial maximum, mid-Holocene, and present. J. Geophys. Res. Atmos. 2003, 108, 108. [Google Scholar] [CrossRef] [Green Version]
- Metcalfe, S.E.; O’Hara, S.L.; Caballero, M.; Davies, S.J. Records of Late Pleistocene–Holocene climatic change in Mexico—A review. Quat. Sci. Rev. 2000, 19, 699–721. [Google Scholar] [CrossRef]
- Mijnsbrugge, K.V.; Bischoff, A.; Smith, B. A question of origin: Where and how to collect seed for ecological restoration. Basic Appl. Ecol. 2010, 11, 300–311. [Google Scholar] [CrossRef] [Green Version]
- McKay, J.K.; Christian, C.E.; Harrison, S.; Rice, K.J. “How Local Is Local?”—A Review of Practical and Conceptual Issues in the Genetics of Restoration. Restor. Ecol. 2005, 13, 432–440. [Google Scholar] [CrossRef]
- Seager, R.; Vecchi, G. Greenhouse warming and the 21st century hydroclimate of southwestern North America. Proc. Natl. Acad. Sci. USA 2010, 107, 21277–21282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, A. Drought under global warming: A review. Clim. Change 2011, 2, 45–65. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.; Doyle, J. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van De Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Whitlock, M.C.; Mccauley, D.E. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm + 1). Heredity 1999, 82, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Gent, P.R.; Danabasoglu, G.; Donner, L.J.; Holland, M.M.; Hunke, E.C.; Jayne, S.R.; Lawrence, D.M.; Neale, R.B.; Rasch, P.J.; Vertenstein, M.; et al. The Community Climate System Model Version 4. J. Clim. 2011, 24, 4973–4991. [Google Scholar] [CrossRef]
- Hajima, T.; Abe, M.; Arakawa, O.; Suzuki, T.; Komuro, Y.; Ogura, T.; Ogochi, K.; Watanabe, M.; Yamamoto, A.; Tatebe, H.; et al. MIROC MIROC-ES2L Model Output Prepared for CMIP6 CMIP Historical. Available online: https://cera-www.dkrz.de/WDCC/ui/cerasearch/cmip6?input=CMIP6.CMIP.MIROC.MIROC-ES2L.historical (accessed on 6 January 2022).
- Thomson, A.M.; Calvin, K.V.; Smith, S.J.; Kyle, G.P.; Volke, A.; Patel, P.; Delgado-Arias, S.; Bond-Lamberty, B.; Wise, M.A.; Clarke, L.E.; et al. RCP4.5: A pathway for stabilization of radiative forcing by 2100. Clim. Change 2011, 109, 77. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Valverde, A. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Glob. Ecol. Biogeogr. 2012, 21, 498–507. [Google Scholar] [CrossRef]
- Raes, N.; ter Steege, H. A null-model for significance testing of presence-only species distribution models. Ecography 2007, 30, 727–736. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]






| Genetic Cluster | Percentage of Polymorphic Loci | Average Number of Alleles per Locus | Average Effective Number of Alleles per Locus | I | He |
|---|---|---|---|---|---|
| Cluster I | 63.4 | 1.59 b | 1.38 b | 0.333 b | 0.223 b |
| Cluster II | 66.6 | 1.61 b | 1.37 b | 0.337 b | 0.224 b |
| Cluster III | 73.1 | 1.67 a | 1.45 a | 0.389 a | 0.261 a |
| Average | 67.4 | 1.62 | 1.40 | 0.353 | 0.236 |
| ID | Variable | Cluster | ||
|---|---|---|---|---|
| I | II | III | ||
| Bio2 | Mean diurnal range | 1.4 | 18.1 | 2.3 |
| Bio4 | Temperature seasonality | 2.0 | 11.5 | 0 |
| Bio9 | Mean temperature of the driest quarter | 2.0 | 0.0 | 57.7 |
| Bio10 | Mean temperature of the warmest quarter | 35.2 | 45.0 | 4.7 |
| Bio11 | Mean temperature of the coldest quarter | 0.6 | 0.0 | 1.9 |
| Bio15 | Precipitation seasonality | 44.0 | 25.4 | 8.4 |
| Bio17 | Precipitation of the driest quarter | 14.7 | 0.0 | 25 |
| Environmental Niche Model | Genetic Cluster | Total | ||
|---|---|---|---|---|
| Cluster I (km−2) | Cluster II (km−2) | Cluster III (km−2) | ||
| Last inter-glacial (120,000–140,000 years ago) | 13,257.3 | 22,962.2 | 14,789.5 | 53,009.00 |
| Last glacial maximum (21,000 years ago) | 18,506.70 | 22,887.1 | 17,668.4 | 54,062.2 |
| Present prediction (1970–2000) | 15,659.7 | 16,961.8 | 8107.6 | 40,729.2 |
| Near future (2021–2040) | 10,449.1 | 22,152.78 | 7968.9 | 40,570.7 |
| Mid-century (2041–2060) | 12,145.8 | 21,361.1 | 17,888.8 | 51,395.7 |
| Environmental Niche Model | Cluster I vs. II | Cluster I vs. III | Cluster II vs. III | |||
|---|---|---|---|---|---|---|
| SD | WI | SD | WI | SD | WI | |
| Last inter-glacial (120,000–140,000 years ago) | 0.47 * | 0.73 * | 0.44 * | 0.73 * | 0.18 * | 0.39 * |
| Last glacial maximum (21,000 years ago) | 0.43 * | 0.68 * | 0.48 * | 0.76 * | 0.17 * | 0.35 * |
| Present prediction (1970–2000) | 0.43 * | 0.68 * | 0.42 * | 0.69 * | 0.18 * | 0.33 * |
| Near future (2021–2040) | 0.41 * | 0.61 * | 0.47 * | 0.72 * | 0.30 * | 0.47 * |
| Mid-century (2041–2060) | 0.43 * | 0.72 * | 0.24 * | 0.51 * | 0.13 * | 0.25 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Nieto, C.R.; Villarreal-Guerrero, F.; Jurado-Guerra, P.; Ochoa-Rivero, J.M.; Prieto-Amparán, J.A.; Corrales-Lerma, R.; Pinedo-Alvarez, A.; Álvarez-Holguín, A. Environmental Niche Dynamics of Blue Grama (Bouteloua gracilis) Ecotypes in Northern Mexico: Genetic Structure and Implications for Restoration Management. Plants 2022, 11, 684. https://doi.org/10.3390/plants11050684
Morales-Nieto CR, Villarreal-Guerrero F, Jurado-Guerra P, Ochoa-Rivero JM, Prieto-Amparán JA, Corrales-Lerma R, Pinedo-Alvarez A, Álvarez-Holguín A. Environmental Niche Dynamics of Blue Grama (Bouteloua gracilis) Ecotypes in Northern Mexico: Genetic Structure and Implications for Restoration Management. Plants. 2022; 11(5):684. https://doi.org/10.3390/plants11050684
Chicago/Turabian StyleMorales-Nieto, Carlos R., Federico Villarreal-Guerrero, Pedro Jurado-Guerra, Jesús M. Ochoa-Rivero, Jesús A. Prieto-Amparán, Raúl Corrales-Lerma, Alfredo Pinedo-Alvarez, and Alan Álvarez-Holguín. 2022. "Environmental Niche Dynamics of Blue Grama (Bouteloua gracilis) Ecotypes in Northern Mexico: Genetic Structure and Implications for Restoration Management" Plants 11, no. 5: 684. https://doi.org/10.3390/plants11050684