DNA Barcoding Study of Representative Thymus Species in Bulgaria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Efficiency of PCR Amplification and Sequencing
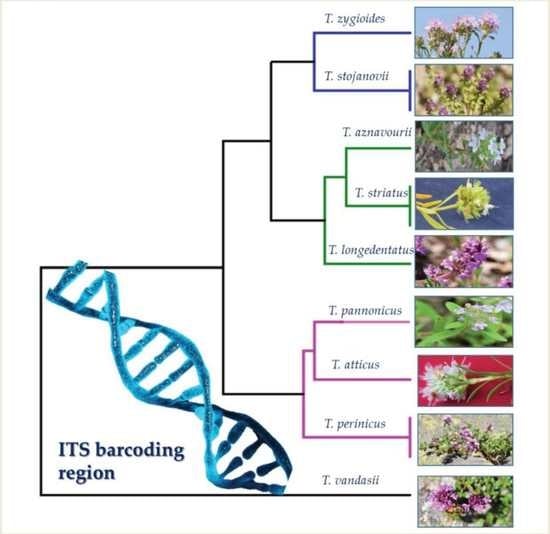
2.2. Genetic Diversity of Thymus Species and Accessions
3. Material and Methods
3.1. Plant Material
3.2. DNA Extraction, PCR Amplification and Sequencing
3.3. Sequence Alignment and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Federici, S.; Galimberti, A.; Bartolucci, F.; Bruni, I.; De Mattia, F.; Cortis, P.; Labra, M. DNA barcoding to analyse taxonomically complex groups in plants: The case of Thymus (Lamiaceae). Bot. J. Linn. Soc. 2013, 171, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Jalas, J. Notes on Thymus L. (Labiatae) in Europe. I. Supraspecific classification and nomenclature. Bot. J. Linn. Soc. 1971, 64, 199–215. [Google Scholar] [CrossRef]
- Jalas, J. Thymus. In Flora Europaea; Tutin, T., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Eds.; University Press: Cambridge, UK, 1972; Volume 3, pp. 172–182. [Google Scholar]
- Baden, C. Thymus L. In Mountain Flora of Greece; Strid, A., Tan, K., Eds.; Edinburgh University Press: Edinburgh, UK, 1986; pp. 139–165. [Google Scholar]
- Morales, R. The history, botany and taxonomy of genus Thymus. In Thyme. The Genus Thymus; Stahl-Biskup, E., Sáez, F., Eds.; Taylor & Francis: London, UK; New York, NY, USA, 2002; pp. 1–43. [Google Scholar]
- Bartolucci, F. Verso una revisione biosistematatica del genere Thymus L. In Italia: Considerazioni Nomenclaturali, Sistematiche e Criticità Tassonomica; Annali di Botanica: Rome, Italy, 2009; Volume 2010, pp. 135–148, Supplemento. [Google Scholar]
- Starr, J.R.; Naczi, R.F.C.; Chouinard, B.N. Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae). Mol. Ecol. Resour. 2009, 9 (Suppl. 1), 151–163. [Google Scholar] [CrossRef] [PubMed]
- Markova, M. Thymus L. In Flora of PR Bulgaria; Velchev, V., Ed.; BAS: Sofia, Bulgaria, 1989; Volume 9, pp. 288–332. (In Bulgarian) [Google Scholar]
- Pavlova, D.; Kozuharova, E.K.; Dimitrov, D.S. The serpentine flora in the Eastern Rhodopes Mountains (Bulgaria). In Progress in Botanical Research: Proceedings of the 1st Balkan Botanical Congress; Tsekos, I., Moustakas, M., Eds.; Kluwer Acdemic Publishers: Dordrecht, The Netherlands, 1998; pp. 133–136. [Google Scholar]
- Stoyanov, S.; Marinov, Y. Thymus jalasianus (Lamiaceae), a new species from the serpentine area of the Eastern Rhodope Mountains, Bulgaria. Ann. Bot. Fenn. 2020, 57, 163–172. [Google Scholar] [CrossRef]
- Stoyanov, S.; Marinov, Y. Thymus aznavourii (Lamiaceae): First records for Bulgarian and Greek flora. Compt. Rend. Acad. Bulg. Sci. Sci. Math. Nat. 2021, 74, 352–362. [Google Scholar] [CrossRef]
- Aneva, I.; Zhelev, P.; Stoyanov, S.; Marinov, Y.; Georgieva, K. Survey on the distribution, diversity and phytochemistry of genus Thymus in Bulgaria. Ecol. Balk. 2018, 10, 101–110. [Google Scholar]
- Aneva, I.; Trendafilova, A.; Nikolova, M.; Todorova, M.; Georgieva, K. Essential oil composition of the Balkan endemic Thymus longedentatus (Degen & Urum.) Ronniger. BLACPMA 2019, 18, 197–203. [Google Scholar] [CrossRef]
- Trendafilova, A.; Todorova, M.; Ivanova, V.; Zhelev, P.; Aneva, I. Essential oil composition of five Thymus species from Bulgaria. Chem. Biodivers. 2021, 18, e2100498. [Google Scholar] [CrossRef] [PubMed]
- Sostarić, I.; Liber, Z.; Grdiša, M.; Marin, P.D.; Stevanović, Z.; Satović, Z. Genetic diversity and relationships among species of the genus Thymus L. (section Serpyllum). Flora 2012, 207, 654–661. [Google Scholar] [CrossRef]
- Kulevanova, S.; Stoeva, T.; Ristić, M. The essential oil composition of Thymus tosevii and Thymus macedonicus from Bulgaria. Boll. Chim. Farm. 2000, 139, 85–88. [Google Scholar]
- Marin, P.D.; Grayer, R.J.; Kite, G.C.; Matevski, V. External leaf flavonoids of Thymus species from Macedonia. Biochem. Syst. Ecol. 2003, 31, 1291–1307. [Google Scholar] [CrossRef]
- Damianova, S.; Tasheva, S.; Stoyanova, A.; Damianov, D. Investigation of extracts from Thyme (Thymus vulgaris L.) for application in cosmetics. J. Essent. Oil Bear. Plants 2008, 11, 443–450. [Google Scholar] [CrossRef]
- Vidić, D.; Ćavar, S.; Solić, M.E.; Maksimović, M. Volatile constituents of two rare subspecies of Thymus praecox. Nat. Prod. Commun. 2010, 5, 1123–1126. [Google Scholar] [CrossRef] [Green Version]
- Željković, S.Ć.; Maksimović, M. Chemical composition and bioactivity of essential oil from Thymus species in Balkan Peninsula. Phytochem. Rev. 2003, 14, 335–352. [Google Scholar] [CrossRef]
- Tarayre, M.; Saumitou-Laprade, P.; Cuguenl, J.; Couvet, D.; Thompson, J. The spatial genetic structure of cytoplasmic (cpDNA) and nuclear (allozyme) markers within and among populations of the gynodioecious Thymus vulgaris (Labiatae) in southern France. Am. J. Bot. 1997, 84, 1675–1684. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D. Population structure and the spatial dynamics of genetic polymorphism in thyme. In Thyme. The Genus Thymus; Stahl-Biskup, E., Sáez, F., Eds.; Taylor & Francis: London, UK, 2002; pp. 44–74. [Google Scholar]
- Soorni, A.; Borna, T.; Alemardan, A.; Chakrabarti, M.; Hunt, A.G.; Bombarely, A. Transcriptome landscape variation in the genus Thymus. Genes 2019, 10, 620. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Erickson, D.L. (Eds.) DNA Barcodes: Methods and Protocols; Humana Press, Springer ScienceþPublishing Media, LLC: New York, NY, USA, 2012; p. 470. [Google Scholar]
- Bräuchler, C.; Meimberg, H.; Heubl, G. Molecular phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae)—Taxonomy, biogeography and conflicts. Mol. Phylogenet. Evol. 2010, 55, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Chen, Y.-P.; Salmaki, Y.; Drew, B.T.; Wilson, T.C.; Scheen, A.-C.; Celep, F.; Bräuchler, C.; Bendiksby, M.; Wang, Q.; et al. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 2021, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Drew, B.T.; Sytsma, K.J. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 2012, 99, 933–953. [Google Scholar] [CrossRef] [Green Version]
- De Mattia, F.; Bruni, I.; Galimberti, A.; Cattaneo, F.; Casiraghi, M.; Labra, M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiaceae. Food Res. Int. 2011, 44, 693–702. [Google Scholar] [CrossRef]
- Theodoridis, S.; Stefanaki, A.; Tezcan, M.; Aki, C.; Kokkini, S.; Vlachonasios, K.E. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çesme-Karaburun Peninsula (Turkey). Mol. Ecol. Resour. 2012, 12, 620–633. [Google Scholar] [CrossRef]
- Abdolahinia, E.D.; Bashir, N.S.; Hagnazari, A.; Nazemiyeh, H.; Hejazi, M.S. A Comparative phenotypic and ITS based genotypic study in Thyme species (Thymus L. Lamiaceae). Int. J. Plant Res. 2011, 24, 102–113. [Google Scholar]
- Sonboli, A.; Mirjalili, M.H.; Bakhtiar, Z.; Jamzad, Z. Molecular authentication of Thymus persicus based on nrDNA ITS sequence data. Iran J. Bot. 2013, 19, 179–185. [Google Scholar]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 206, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Lahaye, R.; van der Bank, M.; Bogarin, D.; Warner, J.; Pupulin, F.; Gigot, G.; Maurin, O.; Duthoit, S.; Barraclough, T.G.; Savolainen, V. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA 2008, 105, 2923–2928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newmaster, S.G.; Fazekas, A.J.; Steeves, A.D.; Janovec, J. Testing candidate plant barcode regions in the Myristicaceae. Mol. Ecol. Resour. 2008, 8, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Muellner, A.M.; Schaefer, H.; Lahaye, R. Evaluation of candidate DNA barcoding loci for economically important timber species of the mahogany family (Meliaceae). Mol. Ecol. Resour. 2011, 11, 450–460. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the noncoding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingsworth, M.L.; Clark, A.A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Wendel, J. Plant speciation—Rise of the poor cousins. New Phytol. 2004, 161, 1–21. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Wood, T.E.; Baack, E.J. The nature of plant species. Nature 2006, 440, 524–527. [Google Scholar] [CrossRef]
- Ennos, R.A.; French, G.C.; Hollingsworth, P.M. Conserving taxonomic complexity. Trends Ecol. Evol. 2005, 20, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the Human-Ape Splitting by a molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]


| Barcode Region | N (Samples Tested) | Alignment Length (bp) | Percentage of Amplification Efficiency | Percentage of Sequencing Efficiency (from Amplified Barcodes) |
|---|---|---|---|---|
| matK | 14 | 760 | 100 | 100 |
| rbcL | 14 | 530 | 100 | 100 |
| trnH-psbA | 14 | 350 | 100 | 100 |
| ITS | 14 | 619 | 93.4 | 93.4 |
| DNA Barcode Region | Ns | C | V | Pi | S | Average Pairwise Distance/SE |
|---|---|---|---|---|---|---|
| rbcL | 529 | 527 | 2 | 2 | 0 | 0.00127/0.00009 |
| trnH-psbA | 351 | 333 | 16 | 10 | 6 | 0.13205/0.00371 |
| matK | 761 | 748 | 13 | 3 | 10 | 0.00342/0.00171 |
| ITS | 618 | 604 | 14 | 2 | 12 | 0.00464/0.00144 |
| rbcL+matK | 1290 | 1275 | 15 | 5 | 10 | 0.00230/0.00066 |
| rbcL+trnH-psbA | 880 | 860 | 18 | 12 | 6 | 0.00577/0.00147 |
| matK+trnH-psbA | 1112 | 1081 | 29 | 13 | 16 | 0.00748/0.00176 |
| Species | Sample Code | Geographic Coordinates Altitude |
|---|---|---|
| Section Hyphodromi (A. Kerner) Halácsy | ||
| Subsection Subbracteati (Klokov) Jalas | ||
| Thymus atticus Čelak. | T08 | 41°25′46″ N 23°42′42″ E 760 m |
| Thymus perinicus (Velen.) Jalas | T57 | 41°46′13″ N 23°24′28″ E |
| T58 | 2450 m | |
| Thymus striatus Vahl. | T28 | 41°57′52″ N 22°56′10″ E 900 m |
| Subsection Serpyllastrum Huguet del Villar | ||
| Thymus aznavourii Velen. | T69 | 41°50′04″ N 26°28′55″ E 120 m |
| Thymus zygioides Griseb. | T26 | 41°39′07″ N 25°50′39″ E 300 m |
| Section Serpyllum (Miller) Bentham | ||
| Subsection Isolepides (Borbás) Halácsy | ||
| Thymus longedentatus (Deg. et Urum.) Ronniger | T17 | 42° 03′24″ N 24°26′17″ E 300 m |
| T27 | 41°40′30″ N 25°49′57″ E 480 m | |
| Thymus pannonicus All. | T25 | 41°39′06″ N 25°50′39″ E 300 m |
| Subsection Alternantes Klokov | ||
| Thymus pulegioides L. | T38 | 41°47′ 07″ N 23°27′40″ E 1500 m |
| Subsection Pseudomarginati (Braun ex Borbás) Jalas | ||
| Thymus stojanovii Deg. | T52 | 41°24′37″ N 23°38′57″ E 1600 m |
| T56 | 41°33′04″ N 24°25′46″ E 1300 m | |
| Thymus thracicus Velen. | T14 | 41°46′14″ N 23°24′48″ E 2200 m |
| Thymus vandasii Velen. | T62 | 42°12′18″ N 23°19′20″ E 2200 m |
| Barcode Region | Primers | Primer Sequences 5′-3′ | PCR Conditions |
|---|---|---|---|
| matK | MatK-RKIM-f | ACCCAGTCCATCTGGAAATCTTGGTTC | 95 °C 5 min 95 °C 30 s, 51 °C 50 s 72 °C 1.4 min, 35 cycles 72 °C 7 min |
| MatK-3FKIM-r | CGTACAGTACTTTTGTGTTTACGAG | ||
| rbcL | rbcLa-F | ATGTCACCACAAACAGAGACTAAAGC | 94 °C 4 min 94 °C 30 s 55 °C 30 s 72 °C 1 min, 35 cycles 72 °C 10 min |
| rbcLajf634R | GAAACGGTCTCTCCAACGCAT | ||
| trnH-psbA | psbA-trnH | CGCGCATGGTGGATTCACAATCC | 94 °C 4 min 94 °C 30 s, 55 °C 30 s, 72 °C 1 min, 35 cycles 72 °C 7 min |
| psbA-3F | GTTATGCATGAACGTAATGCTC | ||
| ITS | ITS_F1 | CCTTATCATTTAGAGGAAGGAG | 94 °C 5 min 94 °C 30 s, 50 °C 30 s, 72 °C 1min, 35 cycles 72 °C 5 min |
| ITS 4 | TCCTCCGCTTATTGATATGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aneva, I.; Zhelev, P.; Bonchev, G.; Boycheva, I.; Simeonova, S.; Kancheva, D. DNA Barcoding Study of Representative Thymus Species in Bulgaria. Plants 2022, 11, 270. https://doi.org/10.3390/plants11030270
Aneva I, Zhelev P, Bonchev G, Boycheva I, Simeonova S, Kancheva D. DNA Barcoding Study of Representative Thymus Species in Bulgaria. Plants. 2022; 11(3):270. https://doi.org/10.3390/plants11030270
Chicago/Turabian StyleAneva, Ina, Petar Zhelev, Georgi Bonchev, Irina Boycheva, Stiliana Simeonova, and Denitsa Kancheva. 2022. "DNA Barcoding Study of Representative Thymus Species in Bulgaria" Plants 11, no. 3: 270. https://doi.org/10.3390/plants11030270