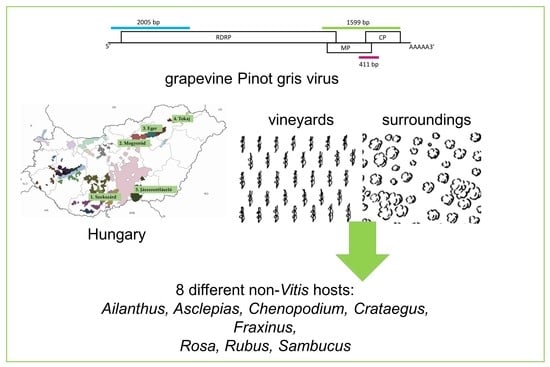
Grapevine Pinot Gris Virus Is Present in Different Non-Vitis Hosts
Abstract
:1. Introduction
2. Results
2.1. GPGV Is Present in Plants Species Other Than Grapevines
2.2. GPGV Variants Are Very Similar and Cluster According to the Sample Location
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. Virus Diagnostics via RT-PCR
4.3. Virus Detection via Northern Blot
4.4. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small rnas in cv pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef]
- Glasa, M.; Predajňa, L.; Komínek, P.; Nagyová, A.; Candresse, T.; Olmos, A. Molecular characterization of divergent grapevine pinot gris virus isolates and their detection in slovak and czech grapevines. Arch. Virol. 2014, 159, 2103–2107. [Google Scholar]
- Eichmeier, A.; Hakalova, E.; Pavelková, R.; Mynarzová, Z.; Saldarelli, P. Detection of grapevine pinot gris virus in certified grapevine stocks in moravia, czech republic. J. Plant Pathol. 2016, 98, 155–157. [Google Scholar] [CrossRef]
- Czotter, N.; Molnar, J.; Szabó, E.; Demian, E.; Kontra, L.; Baksa, I.; Szittya, G.; Kocsis, L.; Deak, T.; Bisztray, G.; et al. NGS of Virus-Derived Small RNAs as a Diagnostic Method Used to Determine Viromes of Hungarian Vineyards. Front. Microbiol. 2018, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Demian, E.; Jaksa-Czotter, N.; Molnar, J.; Tusnady, G.E.; Kocsis, L.; Varallyay, E. Grapevine rootstocks can be a source of infection with non-regulated viruses. Eur. J. Plant Pathol. 2020, 156, 897–912. [Google Scholar]
- Bertazzon, N.; Filippin, L.; Forte, V.; Angelini, E. Grapevine Pinot gris virus seems to have recently been introduced to vineyards in Veneto, Italy. Arch. Virol. 2016, 161, 711–714. [Google Scholar] [CrossRef]
- Cieniewicz, E.J.; Qiu, W.; Saldarelli, P.; Fuchs, M. Believing is seeing: Lessons from emerging viruses in grapevine. J. Plant Pathol. 2020, 102, 619–632. [Google Scholar]
- Hily, J.-M.; Poulicard, N.; Candresse, T.; Vigne, E.; Beuve, M.; Renault, L.; Velt, A.; Spilmont, A.-S.; Lemaire, O. Datamining, genetic diversity analyses, and phylogeographic reconstructions redefine the worldwide evolutionary history of grapevine pinot gris virus and grapevine berry inner necrosis virus. Phytobiomes J. 2019, 4, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Saldarelli, P.; Giampetruzzi, A.; Morelli, M.; Malossini, U.; Pirolo, C.; Bianchedi, P.; Gualandri, V. Genetic Variability of Grapevine Pinot gris virus and Its Association with Grapevine Leaf Mottling and Deformation. Phytopathology 2015, 105, 555–563. [Google Scholar] [CrossRef] [Green Version]
- Malagnini, V.; Duso, C.; Valenzano, D.; Pozzebon, A.; Simonetti, L.; Bianchedi, P.; Saldarelli, P.; Abou Kubaa, R.; de Lillo, E.; Gualandri, V. Role of colomerus vitis (pagenstecher) in the epidemiology of grapevine leaf mottling and deformation in north-eastern italy. In Proceedings of the 19th Conference of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG), Santiago, Chile, 9–12 April 2018; pp. 22–23. [Google Scholar]
- Gualandri, V.; Asquini, E.; Bianchedi, P.; Covelli, L.; Brilli, M.; Malossini, U.; Bragagna, P.; Saldarelli, P.; Si-Ammour, A. Identification of herbaceous hosts of the grapevine pinot gris virus (GPGV). Eur. J. Plant Pathol. 2017, 147, 21–25. [Google Scholar]
- Tarquini, G.; De Amicis, F.; Martini, M.; Ermacora, P.; Loi, N.; Musetti, R.; Bianchi, G.L.; Firrao, G. Analysis of new grapevine pinot gris virus (gpgv) isolates from northeast italy provides clues to track the evolution of a newly emerging clade. Arch. Virol. 2019, 164, 1655–1660. [Google Scholar]
- Bertazzon, N.; Forte, V.; Filippin, L.; Causin, R.; Maixner, M.; Angelini, E. Association between genetic variability and titre of grapevine pinot gris virus with disease symptoms. Plant Pathol. 2017, 66, 949–959. [Google Scholar] [CrossRef]
- Massart, S.; Candresse, T.; Gil, J.F.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.-S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A Framework for the Evaluation of Biosecurity, Commercial, Regulatory, and Scientific Impacts of Plant Viruses and Viroids Identified by NGS Technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Demián, E.; Czotter, N.; Várallyay, É. Detection of grapevine pinot gris virus in different non-vitis hosts in hungary. In Proceedings of the 19th Congress of the International Council for the Study of Virus and Virus-Like Diseases of the Grapevine (ICVG), Santiago, Chile, 9–12 April 2018; pp. 24–25. [Google Scholar]
- Morán, F.; Olmos, A.; Lotos, L.; Predajňa, L.; Katis, N.; Glasa, M.; Maliogka, V.; Ruiz-García, A.B. A novel specific duplex real-time rt-pcr method for absolute quantitation of grapevine pinot gris virus in plant material and single mites. PLoS ONE 2018, 13, e0197237. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for rna extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar]
- White, J.L.; Kaper, J. A simple method for detection of viral satellite RNAs in small plant tissue samples. J. Virol. Methods 1989, 23, 83–93. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]





| Sample Collection Site | Host/Cultivar | Date of Sampling | Strain | GPGV Amplified Region | |
|---|---|---|---|---|---|
| 5′ UTR and RdRp Region | 3′ UTR and MP-CP Region | ||||
| Szekszard | grapevine/Kadarka | May 2015 | Sz_Vitis | ON360679 | ON360686 |
| Chenopodium album | Sz_Calbum | ¯ | ON360687 * | ||
| Mogyorod | grapevine/Furmint | June 2016 | M_Vitis | ON360680 | ON360688 |
| Asclepias syriaca | M_Asyr | ON360681 | ON360689 | ||
| Ailanthus sp. | M_Ailanthus | ¯ | ON360690 | ||
| Rosa canina | M_Rosa | ¯ | ON360691 | ||
| Crataegus sp. | M_Crataegus | ¯ | ON360692 | ||
| Sambucus sp. | M_Sambucus | ¯ | ON360693 | ||
| Eger | grapevine/Kadarka | May 2016 | E_Vitis | ON360682 | ON360694 |
| Rosa canina | E_Rosa | ON360683 | ON360695 | ||
| Tokaj | grapevine/Furmint | June 2015 | T_Vitis | ON360684 | ON360696 |
| Rubus sp. | T_Rubus | ON360685 | ON360697 | ||
| Jaszszentlaszlo | grapevine | August 2017 | J_Vitis | ¯ | ON360698 |
| Fraxinus sp. | J_Fraxinus | ¯ | ON360699 | ||
| Similarity to Reference | |||||||
|---|---|---|---|---|---|---|---|
| Similarity to NC_015782.2_Vitis | Similarity to KU312039_Silene | ||||||
| Strain | GB Accession | % nt Level | % aa Level | Strain | GB Accession | % nt Level | % aa Level |
| Sz_Vitis | ON360679 | 97.16 | 97.66 | ||||
| M_Vitis | ON360680 | 97.21 | 97.34 | M_Asyr | ON360681 | 97.36 | 98.28 |
| E_Vitis | ON360682 | 97.91 | 98.28 | E_Rosa | ON360683 | 97.91 | 97.66 |
| T_Vitis | ON360684 | 97.16 | 97.19 | T_Rubus | ON360685 | 96.86 | 98.44 |
| Similarity to Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Similarity to NC_015782.2_Vitis | Similarity to KU312039_Silene | ||||||||
| Strain | GB Accession | % nt Level | MP % aa Level | CP % aa Level | Strain | GB Accession | % nt Level | MP % aa Level | CP % aa Level |
| Sz_Vitis | ON360686 | 96.94 | 96.21 | 99.49 | Sz_Calbum | ON360687 | 97.57 | 97.08 | |
| M_Vitis | ON360688 | 96.62 | 95.39 | 98.46 | M_Asyr | ON360689 | 97.81 | 98.13 | 97.95 |
| M_Ailanthus | ON360690 | 97.31 | 97.33 | 98.46 | |||||
| M_Rosa | ON360691 | 98.38 | 98.93 | 98.97 | |||||
| M_Crataegus | ON360692 | 97.94 | 98.67 | 98.46 | |||||
| M_Sambucus | ON360693 | 98.38 | 99.47 | 98.97 | |||||
| E_Vitis | ON360694 | 97.06 | 95.66 | 100.00 | E_Rosa | ON360695 | 97.81 | 97.87 | 98.97 |
| T_Vitis | ON360696 | 97.12 | 96.21 | 99.49 | T_Rubus | ON360697 | 97.94 | 98.67 | 98.46 |
| J_Vitis | ON360698 | 96.75 | 95.66 | 99.49 | J_Fraxinus | ON360699 | 98.38 | 99.47 | 98.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demian, E.; Jaksa-Czotter, N.; Varallyay, E. Grapevine Pinot Gris Virus Is Present in Different Non-Vitis Hosts. Plants 2022, 11, 1830. https://doi.org/10.3390/plants11141830
Demian E, Jaksa-Czotter N, Varallyay E. Grapevine Pinot Gris Virus Is Present in Different Non-Vitis Hosts. Plants. 2022; 11(14):1830. https://doi.org/10.3390/plants11141830
Chicago/Turabian StyleDemian, Emese, Nikoletta Jaksa-Czotter, and Eva Varallyay. 2022. "Grapevine Pinot Gris Virus Is Present in Different Non-Vitis Hosts" Plants 11, no. 14: 1830. https://doi.org/10.3390/plants11141830