Identification and Expression Analysis of the Genes Involved in the Raffinose Family Oligosaccharides Pathway of Phaseolus vulgaris and Glycine max
Abstract
:1. Introduction
2. Results
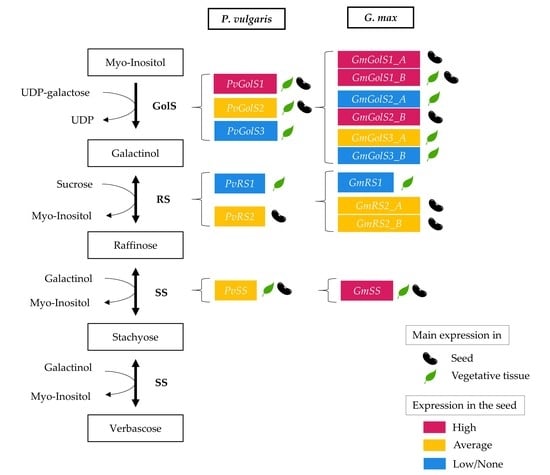
2.1. In Silico Identification of the Galactinol- and RFO Biosynthetic Enzymes in Phaseolus vulgaris and Glycine max
2.2. Transcriptomic Analysis of the RFO Biosynthetic Pathway
2.2.1. Expression Analysis of Galactinol- and RFO Biosynthesis Genes in Glycine max and Phaseolus vulgaris by RNA-seq Re-Analysis
2.2.2. De Novo Expression Analysis of Galactinol- and RFO Biosynthesis Genes during Seed Development in P. vulgaris
2.2.3. Validation of RNA-Sequencing Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. In Silico Identification of the Galactinol and RFO Biosynthetic Genes in Phaseolus vulgaris and Glycine max
4.3. Transcriptome Analysis of the RFO Biosynthesis Pathway
4.3.1. Expression Atlas of Galactinol- and RFO Biosynthesis Genes in G. max and P. vulgaris by RNA-seq Re-Analysis
4.3.2. Expression Atlas of Galactinol- and RFO Biosynthesis Genes during the Seed Development in P. vulgaris by De Novo RNA-seq Analysis
4.4. Validation of RNA-Sequencing Results in P. vulgaris cv. Rosecoco
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Doria, E.; Campion, B.; Sparvoli, F.; Tava, A.; Nielsen, E. Anti-nutrient components and metabolites with health implications in seeds of 10 common bean (Phaseolus vulgaris L. and Phaseolus lunatus L.) landraces cultivated in southern Italy. J. Food Compos. Anal. 2012, 26, 72–80. [Google Scholar] [CrossRef]
- Ganesan, K.; Xu, B. Polyphenol-Rich Dry Common Beans (Phaseolus vulgaris L.) and Their Health Benefits. Int. J. Mol. Sci. 2017, 18, 2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos-Vega, R.; Oomah, B.D.; Loarca-Piña, G.; Vergara-Castañeda, H.A. Common Beans and Their Non-Digestible Fraction: Cancer Inhibitory Activity—An Overview. Foods 2013, 2, 374–392. [Google Scholar] [CrossRef]
- Keefe, S.O.; Bianchi, L.; Sharman, J. Soybean Nutrition. SM J. Nutr. Metab. 2015, 1, 1–9. [Google Scholar]
- Maphosa, Y.; Jideani, V. The Role of Legumes in Human Nutrition. In Functional Food—Improve Health through Adequate Food; Chávarri Hueda, M., Ed.; IntechOpen: London, UK, 2017; ISBN 9781626239777. [Google Scholar] [CrossRef] [Green Version]
- John, K.M.; Khan, F.; Luthria, D.L.; Garrett, W.; Natarajan, S. Proteomic analysis of anti-nutritional factors (ANF’s) in soybean seeds as affected by environmental and genetic factors. Food Chem. 2017, 218, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Dierking, E.C.; Bilyeu, K.D. Association of a Soybean Raffinose Synthase Gene with Low Raffinose and Stachyose Seed Phenotype. Plant Genome 2008, 1, 135–145. [Google Scholar] [CrossRef] [Green Version]
- McPhee, K.E.; Zemetra, R.S.; Brown, J.; Myers, J.R. Genetic Analysis of the Raffinose Family Oligosaccharides in Common Bean. J. Am. Soc. Hortic. Sci. 2002, 127, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Moghaddam, S.M.; Brick, M.A.; Echeverria, D.; Thompson, H.J.; Brick, L.A.; Lee, R.; Mamidi, S.; McClean, P.E. Genetic Architecture of Dietary Fiber and Oligosaccharide Content in a Middle American Panel of Edible Dry Bean. Plant Genome 2018, 11, 170074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguishi, C.T.; Sanada, C.T.; Gouvêa, P.M.; Pandey, A.; Woiciechowski, A.L.; Parada, J.L.; Soccol, C.R. Biotechnological process for producing black bean slurry without stachyose. Food Res. Int. 2009, 42, 425–429. [Google Scholar] [CrossRef]
- Voragen, A.G. Technological aspects of functional food-related carbohydrates. Trends Food Sci. Technol. 1998, 9, 328–335. [Google Scholar] [CrossRef]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 2001, 91, 878–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlin, J.; Lowis, C.; Read, N.W. Investigation of normal flatus production in healthy volunteers. Gut 1991, 32, 665–669. [Google Scholar] [CrossRef] [Green Version]
- Valentine, M.F.; De Tar, J.R.; Mookkan, M.; Firman, J.D.; Zhang, Z.J. Silencing of Soybean Raffinose Synthase Gene Reduced Raffinose Family Oligosaccharides and Increased True Metabolizable Energy of Poultry Feed. Front. Plant Sci. 2017, 8, 692. [Google Scholar] [CrossRef] [Green Version]
- Coon, C.N.; Leske, K.L.; Akavanichan, O.; Cheng, T.K. Effect of Oligosaccharide-Free Soybean Meal on True Metabolizable Energy and Fiber Digestion in Adult Roosters. Poult. Sci. 1990, 69, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Turgeon, R. Mechanisms of phloem loading. Curr. Opin. Plant Biol. 2018, 43, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Haritatos, E.; Keller, F.; Turgeon, R. Raffinose oligosaccharide concentrations measured in individual cell and tissue types in Cucumis melo L. leaves: Implications for phloem loading. Planta 1996, 198, 614–622. [Google Scholar] [CrossRef]
- Eom, J.-S.; Choi, S.-B.; Ward, J.; Jeon, J.-S. The mechanism of phloem loading in rice (Oryza sativa). Mol. Cells 2012, 33, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Yadav, U.P.; Ayre, B.G.; Bush, D.R. Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: Assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 2015, 6, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuther, E.; Buchel, K.; Hundertmark, M.; Stitt, M.; Hincha, D.K.; Heyer, A.G. The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett. 2004, 576, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Zuther, E.; Schulz, E.; Childs, L.H.; Hincha, D.K. Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 2012, 35, 1860–1878. [Google Scholar] [CrossRef]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Elsayed, A.I.; Rafudeen, M.S.; Golldack, D. Physiological aspects of raffinose family oligosaccharides in plants: Protection against abiotic stress. Plant Biol. 2014, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Panikulangara, T.J.; Eggers-Schumacher, G.; Wunderlich, M.; Stransky, H.; Schöffl, F. Galactinol synthase1. A Novel Heat Shock Factor Target Gene Responsible for Heat-Induced Synthesis of Raffinose Family Oligosaccharides in Arabidopsis. Plant Physiol. 2004, 136, 3148–3158. [Google Scholar] [CrossRef] [Green Version]
- Peters, S.W. Raffinose Family Oligaosaccharides (RFOs) are Putative Abiotic Stress Protectants: Case Studies on Frost Tolerance and Water Deficit in Ajuga reptans and Arabidopsis thaliana. Ph.D. Thesis, University of Zurich, Zurich, Switzerland, 2010. [Google Scholar]
- Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Galactinol and Raffinose Constitute a Novel Function to Protect Plants from Oxidative Damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef]
- Hincha, D.K.; Zuther, E.; Heyer, A.G. The preservation of liposomes by raffinose family oligosaccharides during drying is mediated by effects on fusion and lipid phase transitions. Biochim. Biophys. Acta BBA Biomembr. 2003, 1612, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Bailly, C.; Audigier, C.; Ladonne, F.; Wagner, M.-H.; Coste, F.; Corbineau, F.; Côme, D. Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. J. Exp. Bot. 2001, 52, 701–708. [Google Scholar] [CrossRef] [Green Version]
- Blackman, S.A.; Obendorf, R.L.; Leopold, A.C. Maturation Proteins and Sugars in Desiccation Tolerance of Developing Soybean Seeds. Plant Physiol. 1992, 100, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Blöchl, A.; Peterbauer, T.; Richter, A. Inhibition of raffinose oligosaccharide breakdown delays germination of pea seeds. J. Plant Physiol. 2007, 164, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Gangola, M.; Jaiswal, S.; Kannan, U.; Gaur, P.; Båga, M.; Chibbar, R.N. Galactinol synthase enzyme activity influences raffinose family oligosaccharides (RFO) accumulation in developing chickpea (Cicer arietinum L.) seeds. Phytochemistry 2016, 125, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Kandler, O.; Hopf, H. Occurrence, Metabolism, and Function of Oligosaccharides; Academic Press: Cambridge, MA, USA, 1980; Volume 3, ISBN 0126754039. [Google Scholar]
- Frias, J.; Bakhsh, A.; Jones, D.A.; Arthur, A.E.; Vidal-Valverde, C.; Rhodes, M.J.C.; Hedley, C.L. Genetic analysis of the raffinose oligosaccharide pathway in lentil seeds. J. Exp. Bot. 1999, 50, 469–476. [Google Scholar] [CrossRef]
- Peterbauer, T.; Richter, A. Biochemistry and physiology of raffinose family oligosaccharides and galactosyl cyclitols in seeds. Seed Sci. Res. 2001, 11, 185–197. [Google Scholar] [CrossRef]
- Blöchl, A.; Peterbauer, T.; Hofmann, J.; Richter, A. Enzymatic breakdown of raffinose oligosaccharides in pea seeds. Planta 2008, 228, 99–110. [Google Scholar] [CrossRef]
- Dierking, E.C.; Bilyeu, K.D. Raffinose and stachyose metabolism are not required for efficient soybean seed germination. J. Plant Physiol. 2009, 166, 1329–1335. [Google Scholar] [CrossRef]
- Peterbauer, T.; Mach, L.; Mucha, J.; Richter, A. Functional expression of a cDNA encoding pea (Pisum sativum L.) raffinose synthase, partial purification of the enzyme from maturing seeds, and steady-state kinetic analysis of raffinose synthesis. Planta 2002, 215, 839–846. [Google Scholar] [CrossRef]
- Peterbauer, T.; Mucha, J.; Mach, L.; Richter, A. Chain elongation of raffinose in pea seeds. Isolation, characterization, and molecular cloning of a multifunctional enzyme catalyzing the synthesis of stachyose and verbascose. J. Biol. Chem. 2002, 277, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Lahuta, L.B.; Goszczyńska, J. Inhibition of raffinose family oligosaccharides and galactosyl pinitols breakdown delays germination of winter vetch (Vicia villosa Roth.) seeds. Acta Soc. Bot. Pol. 2011, 78, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, M.; Keller, F. Metabolism of the Raffinose Family Oligosaccharides in Leaves of Ajuga reptans L. (Inter- and Intracellular Compartmentation). Plant Physiol. 1995, 109, 991–998. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, G.A.; Wilson, C.; Madore, M.A. Root-Zone Salinity Alters Raffinose Oligosaccharide Metabolism and Transport in Coleus. Plant Physiol. 1997, 115, 1267–1276. [Google Scholar] [CrossRef] [Green Version]
- Le, H.; Nguyen, N.H.; Ta, D.T.; Le, T.N.T.; Bui, T.P.; Le, N.T.; Nguyen, C.X.; Rolletschek, H.; Stacey, G.; Stacey, M.G.; et al. CRISPR/Cas9-Mediated Knockout of Galactinol Synthase-Encoding Genes Reduces Raffinose Family Oligosaccharide Levels in Soybean Seeds. Front. Plant Sci. 2020, 11, 2033. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Calestani, D.V. Contributo alla Sistemica: Ombrellifere D’Europa. Webbia 1905, 1, 89–280. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Zhang, Y.; Wang, D.; Liu, Y.; Dirk, L.M.; Goodman, J.; Downie, A.B.; Wang, J.; Wang, G.; Zhao, T. Regulation of Seed Vigor by Manipulation of Raffinose Family Oligosaccharides in Maize and Arabidopsis thaliana. Mol. Plant 2017, 10, 1540–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterbauer, T.; Mucha, J.; Mayer, U.; Popp, M.; Glossl, J.; Richter, A. Stachyose synthesis in seeds of adzuki bean (Vigna angularis): Molecular cloning and functional expression of stachyose synthase. Plant J. 1999, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Oñate, M.; Ángel, H.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016, 17, 32. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.-A.; Peng, D.-L.; et al. Global Dissection of Alternative Splicing in Paleopolyploid Soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [Green Version]
- Fehr, W.R.; Caviness, C.E. Stages of soybean development. Spec. Rep. 1977, 87, 1–12. [Google Scholar]
- Pereira, W.; Bassinello, P.Z.; Brondani, C.; Vianello, R.P. An improved method for RNA extraction from common bean seeds and validation of reference genes for qPCR. Crop Breed. Appl. Biotechnol. 2017, 17, 150–158. [Google Scholar] [CrossRef]
- Qiu, D.; Vuong, T.; Valliyodan, B.; Shi, H.; Guo, B.; Shannon, J.G.; Nguyen, H.T. Identification and characterization of a stachyose synthase gene controlling reduced stachyose content in soybean. Theor. Appl. Genet. 2015, 128, 2167–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, L.; Chen, L.; Ma, H.; Ruan, Y.; Xu, T.; Xu, C.; He, Y.; Qi, M. Molecular cloning and expression of an encoding galactinol synthase gene (AnGolS1) in seedling of Ammopiptanthus nanus. Sci. Rep. 2016, 6, 36113. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Yuan, S.; He, Y.; Fan, J.; Zhou, Y.; Qiu, T.; Lin, X.; Yao, Y.; Liu, J.; Fu, S.; et al. Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz). Agronomy 2018, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Salvi, P.; Kamble, N.U.; Majee, M. Stress-Inducible Galactinol Synthase of Chickpea (CaGolS) is Implicated in Heat and Oxidative Stress Tolerance Through Reducing Stress-Induced Excessive Reactive Oxygen Species Accumulation. Plant Cell Physiol. 2018, 59, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Peters, S.; Egert, A.; Stieger, B.; Keller, F. Functional Identification of Arabidopsis ATSIP2 (At3g57520) as an Alkaline -Galactosidase with a Substrate Specificity for Raffinose and an Apparent Sink-Specific Expression Pattern. Plant Cell Physiol. 2010, 51, 1815–1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Li, T.; Kim, W.-D.; Kitaoka, M.; Yoshida, S.; Nakajima, M.; Kobayashi, H. Characterization of raffinose synthase from rice (Oryza sativa L. var. Nipponbare). Biotechnol. Lett. 2007, 29, 635–640. [Google Scholar] [CrossRef]
- E McClean, P.; Mamidi, S.; McConnell, M.; Chikara, S.; Lee, R. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genom. 2010, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yu, D.; Choi, H.-K.; Kim, R.W. Reconstruction of a composite comparative map composed of ten legume genomes. Genes Genom. 2017, 39, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.A.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.; Budzinski, I.G.; Marur, C.J.; Petkowicz, C.; Pereira, L.F.P.; Vieira, L.G. Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol. Biochem. 2011, 49, 441–448. [Google Scholar] [CrossRef]
- Zhao, T.-Y.; Thacker, R.; Corum, J.W.; Snyder, J.C.; Meeley, R.B.; Obendorf, R.L.; Downie, B. Expression of the maize GALACTINOL SYNTHASE gene family: (I) Expression of two different genes during seed development and germination. Physiol. Plant. 2004, 121, 634–646. [Google Scholar] [CrossRef]
- Lowell, C.A.; Kuo, T.M. Oligosaccharide Metabolism and Accumulation in Developing Soybean Seeds. Crop. Sci. 1989, 29, 459–465. [Google Scholar] [CrossRef]
- Saldivar, X.; Wang, Y.-J.; Chen, P.; Hou, A. Changes in chemical composition during soybean seed development. Food Chem. 2011, 124, 1369–1375. [Google Scholar] [CrossRef]
- Tsaniklidis, G.; Benovias, A.; Delis, C.; Aivalakis, G. Acidic alpha galactosidase during the maturation and cold storage of cherry tomatoes. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Egert, A.; Keller, F.; Peters, S. Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5, At5g40390). BMC Plant Biol. 2013, 13, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangl, R.; Behmüller, R.; Tenhaken, R. Molecular cloning of AtRS4, a seed specific multifunctional RFO synthase/galactosylhydrolase in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Qi, X.; Wang, Z.; Li, P.; Wu, C.; Zhang, H.; Zhao, Y. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhao, T.-Y.; Iii, J.W.C.; Mullen, J.; Meeley, R.B.; Helentjaris, T.; Martín, D.; Downie, B. An alkaline α-galactosidase transcript is present in maize seeds and cultured embryo cells, and accumulates during stress. Seed Sci. Res. 2006, 16, 107–121. [Google Scholar] [CrossRef]
- Carmi, N.; Zhang, G.; Petreikov, M.; Gao, Z.; Eyal, Y.; Granot, D.; Schaffer, A.A. Cloning and functional expression of alkaline alpha-galactosidase from melon fruit: Similarity to plant SIP proteins uncovers a novel family of plant glycosyl hydrolases. Plant J. 2003, 33, 97–106. [Google Scholar] [CrossRef]
- Imaizumi, C.; Tomatsu, H.; Kitazawa, K.; Yoshimi, Y.; Shibano, S.; Kikuchi, K.; Yamaguchi, M.; Kaneko, S.; Tsumuraya, Y.; Kotake, T. Heterologous expression and characterization of an Arabidopsis β-l-arabinopyranosidase and α-d-galactosidases acting on β-l-arabinopyranosyl residues. J. Exp. Bot. 2017, 68, 4651–4661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182. [Google Scholar] [CrossRef]
- Kiekens, R.; de Koning, R.; Toili, E.; Angenon, G. The Hidden Potential of High-Throughput RNA-Seq Re-Analysis, a Case Study for DHDPS, Key Enzyme of the Aspartate-Derived Lysine Biosynthesis Pathway and Its Role in Abiotic and Biotic Stress Responses in Soybean. 2021. Manuscript submitted for publication. [Google Scholar]
- Joshi, N.; Fass, J. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. Available online: https://github.com/najoshi/sickle (accessed on 15 May 2020).
- Dobin, A.; Gingeras, T.R.; Spring, C.; Flores, R.; Sampson, J.; Knight, R.; Chia, N.; Technologies, H.S. Mapping RNA-seq with STAR. Curr. Protoc. Bioinform. 2016, 51, 586–597. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Kolde, R. Package “pheatmap”. R Packag. 2015, 1, 790. [Google Scholar]
- Aranda, P.S.; Lajoie, D.M.; Jorcyk, C.L. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Toili, M.E.M.; de Koning, R.; Kiekens, R.; Wahome, S.; Githiri, S.M.; Angenon, G. A comparative transcriptome analysis of fast and slow-cooking common beans (Phaseolus vulgaris L.) during seed development reveals differentially expressed genes involved in the hard-to-cook defect. 2021. Manuscript submitted for publication. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Species | Gene Name | Accession Number * | Function | Chromo-Some | Start (bp) | End (bp) | Protein Length (AA) | # Exons | # Transcripts |
|---|---|---|---|---|---|---|---|---|---|
| P. vulgaris | PvGolS1 | Phvul.001G215300 | Galactinol synthase | Chr1 | 47,166,933 | 47,168,674 | 339 | 3 | 2 |
| G. max | GmGolS1_A | Glyma.03G222000 | Chr3 | 42,494,623 | 42,497,111 | 339 | 3 | 1 | |
| G. max | GmGolS1_B | Glyma.19G219100 | Chr19 | 47,148,225 | 47,150,373 | 335 | 3 | 1 | |
| P. vulgaris | PvGolS2 | Phvul.001G223700 | Chr1 | 47,870,097 | 47,872,698 | 327 | 4 | 1 | |
| G. max | GmGolS2_A | Glyma.03G229800 | Chr3 | 43,172,457 | 43,175,687 | 331 | 4 | 1 | |
| G. max | GmGolS2_B | Glyma.19G227800 | Chr19 | 47,911,130 | 47,914,214 | 330 | 4 | 2 | |
| P. vulgaris | PvGolS3 | Phvul.007G203400 | Chr7 | 32,610,928 | 32,612,577 | 326 | 4 | 1 | |
| G. max | GmGolS3_A | Glyma.10G145300 | Chr10 | 38,014,453 | 38,016,396 | 328 | 4 | 3 | |
| G. max | GmGolS3_B | Glyma.20G094500 | Chr20 | 33,759,417 | 33,761,555 | 324 | 4 | 1 | |
| P. vulgaris | PvRS1 | Phvul.004G007100 | Raffinose synthase | Chr4 | 519,197 | 523,594 | 763 | 5 | 1 |
| G. max | GmRS1 | Glyma.05G003900 | Chr5 | 307,461 | 312,091 | 758 | 5 | 1 | |
| P. vulgaris | PvRS2 | Phvul.009G175400 | Chr9 | 26,053,801 | 26,057,657 | 777 | 5 | 1 | |
| G. max | GmRS2_A | Glyma.06G179200 | Chr6 | 15,217,419 | 15,223,877 | 810 | 5 | 2 | |
| G. max | GmRS2_B | Glyma.05G040300 | Chr5 | 3,593,378 | 3,598,821 | 782 | 5 | 1 | |
| P. vulgaris | PvSS | Phvul.001G214300 | Stachyose synthase | Chr1 | 47,049,258 | 47,052,441 | 857 | 4 | 2 |
| G. max | GmSS | Glyma.19G217700 | Chr19 | 47,033,812 | 47,037,286 | 860 | 4 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Koning, R.; Kiekens, R.; Toili, M.E.M.; Angenon, G. Identification and Expression Analysis of the Genes Involved in the Raffinose Family Oligosaccharides Pathway of Phaseolus vulgaris and Glycine max. Plants 2021, 10, 1465. https://doi.org/10.3390/plants10071465
de Koning R, Kiekens R, Toili MEM, Angenon G. Identification and Expression Analysis of the Genes Involved in the Raffinose Family Oligosaccharides Pathway of Phaseolus vulgaris and Glycine max. Plants. 2021; 10(7):1465. https://doi.org/10.3390/plants10071465
Chicago/Turabian Stylede Koning, Ramon, Raphaël Kiekens, Mary Esther Muyoka Toili, and Geert Angenon. 2021. "Identification and Expression Analysis of the Genes Involved in the Raffinose Family Oligosaccharides Pathway of Phaseolus vulgaris and Glycine max" Plants 10, no. 7: 1465. https://doi.org/10.3390/plants10071465