Identification of Candidate Gene for Internode Length in Rice to Enhance Resistance to Lodging Using QTL Analysis
Abstract
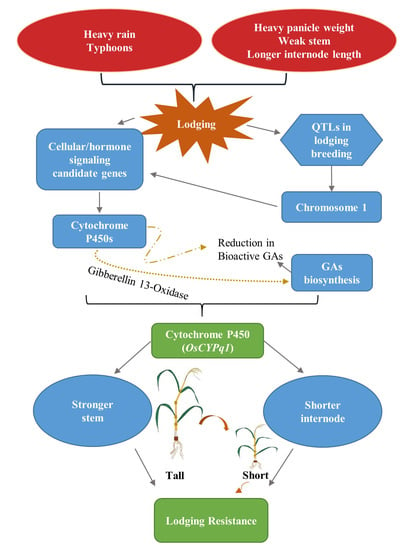
:1. Introduction
2. Results
2.1. Comparison of Agricultural Characteristics and Phenotypic Evaluation
2.2. QTL Analysis Associated with the Length and Stem Diameter of Each Internode
2.3. Based on QTL Mapping Search Candidate Gene
2.4. The Homology Sequence of Candidate Gene and Phylogenetic Tree Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material and Field Experiment Design
4.2. Phenotype Evaluation
4.3. QTL Analysis
4.4. Statistical Analysis and Gene Information Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, D.-D.; Son, J.H.; Lee, G.-S.; Kim, K.-M. Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice. Agronomy 2021, 11, 334. [Google Scholar] [CrossRef]
- Zhang, J.; Li, G.; Song, Y.; Liu, Z.; Yang, C.; Tang, S.; Zheng, C.; Wang, S.; Ding, Y. Lodging resistance characteristics of high-yielding rice populations. Field Crops Res. 2014, 161, 64–74. [Google Scholar] [CrossRef]
- Sowadan, O.; Li, D.; Zhang, Y.; Zhu, S.; Hu, X.; Bhanbhro, L.B.; Edzesi, W.M.; Dang, X.; Hong, D. Mining of favorable alleles for lodging resistance traits in rice (Oryza sativa) through association mapping. Planta 2018, 248, 155–169. [Google Scholar] [CrossRef]
- Long, W.; Dan, D.; Yuan, Z.; Chen, Y.; Jin, J.; Yang, W.; Zhang, Z.; Li, N.; Li, S. Deciphering the Genetic Basis of Lodging Resistance in Wild Rice Oryza longistaminata. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, T.; Madoka, Y.; Hirotsu, N.; Ishimaru, K. Locus prl5 improves lodging resistance of rice by delaying senescence and increasing carbohydrate reaccumulation. Plant Physiol. Biochem. 2006, 44, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Li, Y.; Hu, Z.; Hu, H.; Wang, G.; Li, A.; Wang, Y.; Tu, Y.; Xia, T.; Peng, L.; et al. Ectopic expression of a novel OsExtensin-like gene consistently enhances plant lodging resistance by regulating cell elongation and cell wall thickening in rice. Plant Biotechnol. J. 2018, 16, 254–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Y.; Xu, N.; Wu, Q.; Yu, B.; Li, X.; Chen, R.; Huang, J. Rice transcription factor OsMADS57 regulates plant height by modulating gibberellin catabolism. Rice 2019, 12, 38. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Botwright Acuña, T.; Spielmeyer, W.; Richards, R.A. Effect of gibberellin-sensitive Rht18 and gibberellin-insensitive Rht-D1b dwarfing genes on vegetative and reproductive growth in bread wheat. J. Exp. Bot. 2021, 72, 445–458. [Google Scholar] [CrossRef]
- Shearman, J.R.; Vejchasarn, P.; Naktang, C.; Phansenee, Y.; Jomchai, N.; Lanceras-Siangliw, J.; Tangphatsornruang, S.; Toojinda, T. Rice height QTLs in KDML105 chromosome segment substitution lines. Genomics 2019. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Zhou, Z.; Van Der Straeten, D. Regulation of Submergence-induced Enhanced Shoot Elongation in Oryza sativa L. Ann. Bot. 2003, 91, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Sauter, M.; Mekhedov, S.L.; Kende, H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J. 1995, 7, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J.; Bedinger, P.; Durachko, D.M. Group I allergens of grass pollen as cell wall-loosening agents. Proc. Natl. Acad. Sci. USA 1997, 94, 6559–6564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, L.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Sun, J.; Zhao, H.W.; Yang, L.M.; Zou, D.T. Genetic dissection of rice (Oryza sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 2018, 214, 109. [Google Scholar] [CrossRef]
- Ishimaru, K.; Yano, M.; Aoki, N.; Ono, K.; Hirose, T.; Lin, S.Y.; Monna, L.; Sasaki, T.; Ohsugi, R. Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Genet. 2001, 102, 793–800. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, K.; Murata, K.; Yamazaki, M.; Onosato, K.; Miyao, A.; Hirochika, H. Three Distinct Rice Cellulose Synthase Catalytic Subunit Genes Required for Cellulose Synthesis in the Secondary Wall. Plant Physiol. 2003, 133, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-p.; Li, J.; Deng, X.-W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, T.; Taguchi-Shiobara, F.; Ukai, Y.; Sasaki, T.; Yano, M. Mapping Quantitative Trait Loci for Days-to-heading, and Culm, Panicle and Internode Lengths in a BC1F3 Population Using an Elite Rice Variety, Koshihikari, as the Recurrent Parent. Breed. Sci. 2001, 51, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Mapping of three QTLs that regulate internode elongation in deepwater rice. Breed. Sci. 2008, 58, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A Rice Brassinosteroid-Deficient Mutant, ebisu dwarf (d2), Is Caused by a Loss of Function of a New Member of Cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [Green Version]
- Davidson, S.E.; Reid, J.B.; Helliwell, C.A. Cytochromes P450 in gibberellin biosynthesis. Phytochem. Rev. 2006, 5, 405–419. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Li, Y.; Wei, K. Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol. 2020, 20, 93. [Google Scholar] [CrossRef] [Green Version]
- Hamberger, B.; Ohnishi, T.; Hamberger, B.; Séguin, A.; Bohlmann, J. Evolution of Diterpene Metabolism: Sitka Spruce CYP720B4 Catalyzes Multiple Oxidations in Resin Acid Biosynthesis of Conifer Defense against Insects. Plant Physiol. 2011, 157, 1677–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-España, V.H.; Sánchez-León, N.; Vielle-Calzada, J.-P. CYP85A1 is required for the initiation of female gametogenesis in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 321–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szekeres, M.; Bishop, G.J. 3 Integration of brassinosteroid biosynthesis and signaling. Annu. Plant Rev. Plant Horm. Signal. 2008, 24, 67. [Google Scholar]
- Schuler, M.A. P450s in Plants, Insects, and Their Fungal Pathogens. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Ortiz de Montellano, P.R., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 409–449. [Google Scholar] [CrossRef]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Nomura, T.; Xu, Y.; Zhang, Y.; Peng, Y.; Mao, B.; Hanada, A.; Zhou, H.; Wang, R.; Li, P.; et al. ELONGATED UPPERMOST INTERNODE Encodes a Cytochrome P450 Monooxygenase That Epoxidizes Gibberellins in a Novel Deactivation Reaction in Rice. Plant Cell 2006, 18, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liu, X.; Li, S.; Cheng, Z.; Li, C. The Rice Semi-Dwarf Mutant sd37, Caused by a Mutation in CYP96B4, Plays an Important Role in the Fine-Tuning of Plant Growth. PLoS ONE 2014, 9, e88068. [Google Scholar] [CrossRef]
- Luo, A.; Qian, Q.; Yin, H.; Liu, X.; Yin, C.; Lan, Y.; Tang, J.; Tang, Z.; Cao, S.; Wang, X.; et al. EUI1, Encoding a Putative Cytochrome P450 Monooxygenase, Regulates Internode Elongation by Modulating Gibberellin Responses in Rice. Plant Cell Physiol. 2006, 47, 181–191. [Google Scholar] [CrossRef]
- Wang, H.; Li, W.; Qin, Y.; Pan, Y.; Wang, X.; Weng, Y.; Chen, P.; Li, Y. The Cytochrome P450 Gene CsCYP85A1 Is a Putative Candidate for Super Compact-1 (Scp-1) Plant Architecture Mutation in Cucumber (Cucumis sativus L.). Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Winkler, R.G.; Helentjaris, T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell 1995, 7, 1307–1317. [Google Scholar] [CrossRef] [Green Version]
- Ayano, M.; Kani, T.; Kojima, M.; Sakakibara, H.; Kitaoka, T.; Kuroha, T.; Angeles-Shim, R.B.; Kitano, H.; Nagai, K.; Ashikari, M. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 2014, 37, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, K.; Manigbas, N.L.; Yi, G.; Sohn, J. Identification of quantitative trait loci for resistance to white-backed planthopper (Sogatella furcifera) in rice with Milyang 46 (Cheongcheongbyeo) background. Philipp. J. Crop Sci. 2013, 38, 30–36. [Google Scholar]
- Yun, B.-W.; Kim, M.-G.; Handoyo, T.; Kim, K.-M. Analysis of Rice Grain Quality-Associated Quantitative Trait Loci by Using Genetic Mapping. Am. J. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, S.E.; Daly, M.J.; Lander, E.S. Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0: A tutorial and reference manual. Whitehead Inst. Biomed. Res. Tech. Rep. 1993, 3, 6–40. [Google Scholar]
- McCough, S.R.; Doerge, R.W. QTL mapping in rice. Trends Genet. 1995, 11, 482–487. [Google Scholar] [CrossRef]
- Zeng, Z.B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef] [Green Version]
- Hall, T. BioEdit Version 7.0.0. Distributed by the Author. 2004. Available online: www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 8 May 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Plant Traits | Year | Parents | DH Population | |
|---|---|---|---|---|
| Cheongcheong | Nagdong. | |||
| LP (cm) | 2019 | 21.9 ± 1.0 | 18.6 ± 1.1 | 19.5 ± 2.4 |
| 2020 | 21.5 ± 0.7 | 20.6 ± 2.2 | 19.5 ± 2.5 | |
| LUI (cm) | 2019 | 32.5 ± 1.1 | 30.8 ± 1.3 | 30.1 ± 5.2 |
| 2020 | 31.2 ± 0.5 | 30.8 ± 2.6 | 30.0 ± 5.3 | |
| LSI (cm) | 2019 | 18.6 ± 1.2 | 16.9 ± 1.4 | 16.6 ± 4.2 |
| 2020 | 18.3 ± 0.6 | 15.9 ± 1.1 | 16.7 ± 4.3 | |
| LTI (cm) | 2019 | 13.7 ± 1.1 | 11.4 ± 1.2 | 12.2 ± 3.8 |
| 2020 | 14.8 ± 1.1 | 12.1 ± 1.8 | 12.1 ± 3.7 | |
| LFI (cm) | 2019 | 7.8 ± 1.0 | 4.5 ± 0.2 | 6.8 ± 3.8 |
| 2020 | 9.4 ± 0.6 | 5.3 ± 1.3 | 6.6 ± 3.7 | |
| LLI (cm) | 2019 | 2.8 ± 0.5 | 2.4 ± 0.3 | 2.1 ± 2.4 |
| 2020 | 3.5 ± 0.5 | 2.1 ± 0.3 | 2.0 ± 2.4 | |
| SDUI (mm) | 2019 | 2.5 ± 0.4 | 1.4 ± 0.1 | 2.1 ± 0.45 |
| 2020 | 2.4 ± 0.2 | 1.8 ± 0.3 | 2.1 ± 0.45 | |
| SDSI (mm) | 2019 | 4.5 ± 0.3 | 3.0 ± 0.1 | 3.6 ± 0.6 |
| 2020 | 4.3 ± 0.5 | 3.3 ± 0.3 | 3.5 ± 0.6 | |
| SDTI (mm) | 2019 | 6.0 ± 0.5 | 4.0 ± 0.2 | 4.3 ± 0.7 |
| 2020 | 5.9 ± 0.4 | 4.4 ± 0.4 | 4.3 ± 0.7 | |
| SDFI (mm) | 2019 | 6.6 ± 0.4 | 4.8 ± 0.2 | 4.9 ± 0.7 |
| 2020 | 6.7 ± 0.2 | 4.8 ± 0.4 | 4.9 ± 0.7 | |
| SDLI (mm) | 2019 | 6.8 ± 0.3 | 5.0 ± 0.2 | 4.9 ± 0.7 |
| 2020 | 6.8 ± 0.1 | 5.0 ± 0.6 | 5.1 ± 0.8 | |
| Gene ID | Description |
|---|---|
| Os01g0782200 | Diacylglycerol kinase, catalytic region domain-containing protein. |
| Os01g0783800 | Curculin-like (mannose-binding) lectin domain-containing protein. |
| Os01g0784900 | Basic helix-loop-helix dimerization region bHLH domain-containing protein. |
| Os01g0785400 | GH3 auxin-responsive promoter family protein. |
| Os01g0786900 | WD40-like domain-containing protein. |
| Os01g0794400 | Thioredoxin domain 2 containing protein. |
| Os01g0797600 | Ethylene-responsive element binding factor3 (OsERF3). |
| Os01g0799500 | DNA glycosylase family protein. |
| Os01g0802700 | Auxin Efflux Carrier family protein. |
| Os01g0803900 | Cytochrome P450 family protein. |
| Os01g0823100 | Alpha-expansin OsEXPA2. |
| Os01g0833100 | NLI interacting factor domain-containing protein. |
| Os01g0840300 | Homeobox domain-containing protein. |
| Os01g0851100 | Similar to Axi 1 (Auxin-independent growth promoter)-like protein. |
| Os01g0853400 | Similar to Coronatine-insensitive protein 1. |
| Os01g0854800 | Similar to Cytochrome P450 86A1. |
| Os01g0859500 | Similar to Basic leucine zipper protein (Liguleless2). |
| Os01g0867100 | Aminoacyl-tRNA synthetase, class Ib domain-containing protein. |
| Os01g0868300 | Similar to DNA polymerase alpha catalytic subunit. |
| Os01g0876300 | Galactose oxidase, central domain-containing protein. |
| Os01g0881900 | Leucine-rich repeat, cysteine-containing containing protein. |
| Os01g0883800 | Gibberellin 20 oxidase 2. |
| Os01g0898300 | Armadillo-like helical domain-containing protein. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.-D.; Son, J.-H.; Farooq, M.; Kim, K.-M. Identification of Candidate Gene for Internode Length in Rice to Enhance Resistance to Lodging Using QTL Analysis. Plants 2021, 10, 1369. https://doi.org/10.3390/plants10071369
Zhao D-D, Son J-H, Farooq M, Kim K-M. Identification of Candidate Gene for Internode Length in Rice to Enhance Resistance to Lodging Using QTL Analysis. Plants. 2021; 10(7):1369. https://doi.org/10.3390/plants10071369
Chicago/Turabian StyleZhao, Dan-Dan, Ju-Hyeong Son, Muhammad Farooq, and Kyung-Min Kim. 2021. "Identification of Candidate Gene for Internode Length in Rice to Enhance Resistance to Lodging Using QTL Analysis" Plants 10, no. 7: 1369. https://doi.org/10.3390/plants10071369