Elucidation of the Origin of the Monumental Olive Tree of Vouves in Crete, Greece
Abstract
:1. Introduction
2. Results and Discussion
2.1. Resequencing, Mapping and Variant Calling with the Olea europaea var. sylvestris Reference Genome
2.2. Origin of the Vouves Monumental Olive Tree in the Context of Olive Domestication
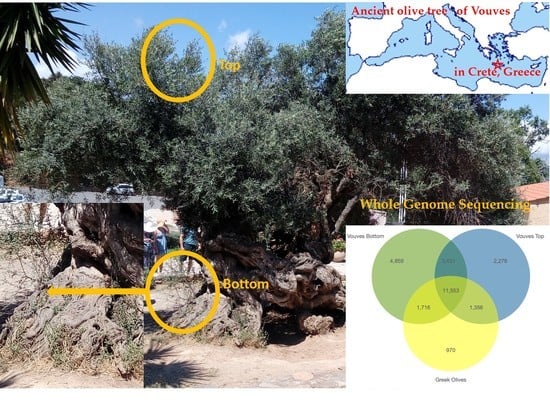
2.3. Gene Space Variation in the Vouves Monumental Olive Tree
3. Conclusions
4. Materials and Methods
4.1. Plant Samples and DNA Extraction
4.2. DNA Sequencing and SSR Analysis
4.3. Read Processing, Reference Mapping and Variant Calling
4.4. Origin Analysis
4.5. Gene Space Comparison
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AJK | Averaged pairwise relatedness index |
| BED | Browser Extensible Data |
| DAPC | Discriminant Analysis of Principal Components |
| GBS | Genotyping-By-Sequencing |
| GFF | Generic Feature Format |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| HI | High Impact variant |
| LI | Low Impact variant |
| MI | Moderate Impact variant |
| PCA | Principal Component Analysis |
| VCF | Variant Call Format |
| cv(s) | cultivar(s) |
References
- International Olive Council. World Olive Oil Figures. 2018. Available online: http://www.internationaloliveoil.org/estaticos/view/131-world-olive-oil-figures (accessed on 1 August 2021).
- Accardi, G.; Aiello, A.; Gargano, V.; Gambino, C.M.; Caracappa, S.; Marineo, S.; Vesco, G.; Carru, C.; Zinellu, A.; Zarcone, M.; et al. Nutraceutical effects of table green olives: A pilot study with Nocellara del Belice olives. Immun. Ageing 2016, 13, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, K.; Melliou, E.; Li, X.; Pedersen, T.L.; Wang, S.C.; Magiatis, P.; Newman, J.W.; Holt, R.R. Oleocanthal-rich extra virgin olive oil demonstrates acute anti-platelet effects in healthy men in a randomized trial. J. Funct. Food 2017, 36, 84–93. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Statistics Division. 2018. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 4 August 2021).
- Besnard, G.; Terral, J.F.; Cornille, A. On the origins and domestication of the olive: A review and perspectives. Ann. Bot. 2018, 121, 385–403. [Google Scholar] [CrossRef] [Green Version]
- Gros-Balthazard, M.; Besnard, G.; Sarah, G.; Holtz, Y.; Leclercq, J.; Santoni, S.; Wegmann, D.; Glémin, S.; Khadari, B. Evolutionary transcriptomics reveals the origins of olives and the genomic changes associated with their domestication. Plant J. 2019, 100, 143–157. [Google Scholar] [CrossRef]
- Dıez, C.M.; Trujillo, I.; Martinez-Urdiroz, N.; Barranco, D.; Rallo, L.; Marfil, P.; Gaut, B.S. Olive domestication and diversification in the Mediterranean Basin. New Phytol. 2015, 206, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Besnard, G.; El Bakkali, A.; Haouane, H.; Baali-Cherif, D.; Moukhli, A.; Khadari, B. Population genetics of Mediterranean and Saharan olives: Geographic patterns of differentiation and evidence for early generations of admixture. Ann. Bot. 2013, 112, 1293–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breton, C.; Tersac, M.; Berville, A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: Several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J. Biogeogr. 2006, 33, 1916–1928. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Lumaret, R.; Ouazzani, N. Plant genetics: Ancient wild olives in Mediterranean forests. Nature 2001, 413, 700. [Google Scholar] [CrossRef]
- Athanasiadis, N.; Gerasimidis, A.; Panagiotidis, S. The olive tree in the pollen diagrams and its significance from a historical and archaeological point of view (in Greek). In Olive Tree and Olive Oil: Workshop in Kalamata 7–9 May 1993; Politistiko Texhnologiko Idryma ETBA: Athens, Greece, 1996. [Google Scholar]
- Margaritis, E. Distinguishing exploitation, domestication, cultivation and production: The olive in the third millennium Aegean. Antiquity 2013, 87, 746–757. [Google Scholar] [CrossRef]
- Homer. Odyssey; GRIGORI: Athens, Greece, 1997. [Google Scholar]
- Herodotus. The Histories; Govosti Publisher: Athens, Greece, 1995. [Google Scholar]
- Martlew, H. A vegetable stew at Gerani Cave. In Minoans and Mycenaeans: Flavours of Their Time; Tzedakis, Y., Martlew, H., Eds.; Ministry of Culture: Athens, Greece, 1999; p. 80. [Google Scholar]
- Beeston, R.F.; Palatinus, J.; Beck, C.W. Organic residue analysis: Chrysokamino. In Archaeology Meets Science: Biomolecular Investigations in Bronze Age Greece; Tzedakis, Y., Martlew, H., Jones, M.K., Eds.; Oxbow Books: Oxford, UK, 2008; pp. 87–107. [Google Scholar]
- Koh, A.; Betancourt, P. Wine and olive oil from an early Minoan hilltop fort. Medit. Archaeol. Archaeom. 2010, 10, 15–23. [Google Scholar]
- Evans, J.; Garner, V. Organic residue in pottery of the Bronze Age in Greece. In Archaeology Meets Science: Biomolecular Investigations in Bronze Age Greece; Tzedakis, Y., Martlew, H., Jones, M.K., Eds.; Oxbow Books: Oxford, UK, 2008; pp. 125–143. [Google Scholar]
- Beck, C.W.; Stout, E.C.; Workulich, K.M.; Phillips., A.J. Absorbed organic residues in pottery from the Minoan settlement of Pseira, Crete. In Archaeology Meets Science: Biomolecular Investigations in Bronze Age Greece; Tzedakis, Y., Martlew, H., Jones, M.K., Eds.; Oxbow Books: Oxford, UK, 2008; pp. 48–73. [Google Scholar]
- Maravelakis, E.; Bilalis, N.; Mantzorou, I.; Konstantaras, A.; Antoniadis, A. 3D modelling of the oldest olive tree of the world. IJCER 2012, 2, 340–347. [Google Scholar]
- Theophrastus. Enquiry into Plants (Greek: Peri Phyton Historia). Books 1–5; Translated by Hort, A.F., Loeb Classical Library; Harvard University Press: Cambridge, MA, USA, 1916; Volume 1–2, ISBN 0-674-99077-3. [Google Scholar]
- Diez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Anestiadou, K.; Nikoloudakis, N.; Hagidimitriou, M.; Katsiotis, A. Monumental olive trees of Cyprus contributed to the establishment of the contemporary olive germplasm. PLoS ONE 2017, 12, e0187697. [Google Scholar] [CrossRef] [Green Version]
- Barazani, O.; Westberg, E.; Hanin, N.; Dag, A.; Kerem, Z.; Tugendhaft, Y.; Hmidat, M.; Hijawi, T.; Kadereit, J.W. A comparative analysis of genetic variation in rootstocks and scions of old olive trees—A window into the history of olive cultivation practices and past genetic variation. BMC Plant Biol. 2014, 14, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barazani, O.; Keren-Keiserman, A.; Westberg, E.; Hanin, N.; Dag, A.; Ben-Ari, G.; Fragman-Sapir, O.; Tugendhaft, Y.; Kerem, Z.; Kadereit, J.W. Genetic variation of naturally growing olive trees in Israel: From abandoned groves to feral and wild? BMC Plant Biol. 2016, 16, 261. [Google Scholar] [CrossRef] [Green Version]
- Langgut, D.; Cheddadi, R.; Carrión, J.S.; Cavanagh, M.; Colombaroli, D.; Eastwood, W.J.; Greenberg, R.; Litt, T.; Mercuri, A.M.; Miebach, A.; et al. The origin and spread of olive cultivation in the Mediterranean Basin: The fossil pollen evidence. Holocene 2019, 29, 902–922. [Google Scholar] [CrossRef]
- Besnard, G.; Garcia-Verdugo, C.; Rubio De Casas, R.; Treier, U.A.; Galland, N.; Vargas, P. Polyploidy in the olive complex (Olea Europaea): Evidence from flow cytometry and nuclear microsatellite analyses. Ann. Bot. 2008, 101, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Rugini, E.; Pannelli, G.; Ceccarelli, M.; Muganuet, M. Isolation of triploid and tetraploid olive (Olea europaea L.) plants from mixoploid cv.‘Frantoio’ and ‘Leccino’ mutants by in vivo and in vitro selection. Plant Breed. 1996, 115, 23–27. [Google Scholar] [CrossRef]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome sequence of the olive tree, Olea europea. GigaScience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Julca, I.; Marcet-Houben, M.; Cruz, F.; Gómez-Garrido, J.; Gaut, B.S.; Díez, C.M.; Gut, I.G.; Alioto, T.S.; Vargas, P.; Gabaldón, T. Genomic evidence for recurrent genetic admixture during the domestication of Mediterranean olive trees (Olea europaea L.). BMC Biol. 2020, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Ruiz, J.; Ramírez-Tejero, J.A.; Fernández-Pozo, N.; de la O Leyva-Pérez, M.; Yan, H.; de la Rosa, R.; Belaj, A.; Montes, E.; Rodríguez-Ariza, O.; Navarro, F.; et al. Transposon activation is a major driver in the genome evolution of cultivated olive trees (Olea europaea L.). Plant Genome 2020, 13, e20010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, G.; Zhang, J.; Liu, X.; Lin, C.; Xin, H.; Xue, L.; Wang, C. De novo assembly of a new Olea Europaea genome accession using nanopore sequencing. Hortic. Res. 2021, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Niu, E.; Shi, A.; Mou, B. Genetic diversity analysis of olive germplasm (Olea europaea L.) with genotyping-by-sequencing technology. Front. Genet. 2019, 10, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koubouris, G.C.; Avramidou, E.V.; Metzidakis, I.T.; Petrakis, P.V.; Sergentani, C.K.; Doulis, A. Phylogenetic and evolutionary applications of analyzing endocarp morphological characters by classification binary tree and leaves by SSR markers for the characterization of olive germplasm. Tree Genet. Genomes 2019, 15, 26. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fu, J.; Xu, Y.; Zhang, J.; Ren, F.; Zhao, H.; Tian, S.; Guo, W.; Tu, X.; Zhao, J.; et al. Genome re-sequencing reveals the evolutionary history of peach fruit edibility. Nat. Commun. 2018, 9, 5404. [Google Scholar] [CrossRef] [Green Version]
- Talavera, A.; Soorni, A.; Bombarely, A.; Matas, A.J.; Hormaza, J.I. Genome-Wide SNP discovery and genomic characterization in avocado (Persea americana Mill.). Sci. Rep. 2019, 9, 20137. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, R.; Belaj, A.; De La Rosa, R.; Leòn, L.; Brizioli, F.; Baldoni, L.; Mousavi, S. EST–SNP study of Olea Europaea L. uncovers functional polymorphisms between cultivated and wild olives. Genes 2020, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Ninot, A.; Howad, W.; Aranzana, M.J.; Senar, R.; Romero, A.; Mariotti, R.; Baldoni, L.; Belaj, A. Survey of over 4500 monumental olive trees preserved on-farm in the northeast Iberian Peninsula, their genotyping and characterization. Sci. Hortic. 2018, 231, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Huang, J.; Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009, 9, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, A.; Puchta, H. The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 2011, 62, 1565–1579. [Google Scholar] [CrossRef] [Green Version]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef]
- Avramidou, E.V.; Koubouris, G.C.; Petrakis, P.V.; Lambrou, K.K.; Metzidakis, I.T.; Doulis, A.G. Classification binary trees with SSR allelic sizes: Combining regression trees with genetic molecular data in order to characterize genetic diversity between cultivars of Olea Europaea L. Agronomy 2020, 10, 1662. [Google Scholar] [CrossRef]
- Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff Fly 2014, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Sample | Total Variants (M) | Heterozygous Variants/100 bp | Total Variants/100 bp | SNP (M) | InDels (M) | MNP (M) |
|---|---|---|---|---|---|---|
| Vouves bottom 1 | 23.26 | 1.31 | 2.09 | 18.79 | 1.27 | 0.12 |
| Vouves top 1 | 19.54 | 1.42 | 1.76 | 15.73 | 1.09 | 0.09 |
| Kalamon 2 | 29.47 | 1.80 | 2.62 | 19.26 | 1.09 | 0.12 |
| Koroneiki 2 | 34.38 | 2.07 | 3.07 | 22.38 | 1.26 | 0.19 |
| Mastoidis 2 | 29.70 | 1.76 | 2.65 | 19.40 | 1.09 | 0.17 |
| Mavreya 2 | 31.80 | 1.99 | 2.84 | 21.32 | 1.18 | 0.12 |
| Megaritiki 2 | 35.70 | 2.16 | 3.20 | 23.24 | 1.30 | 0.18 |
| Myrtolia 2 | 29.10 | 1.69 | 2.60 | 18.75 | 1.08 | 0.16 |
| Sample | % Intergenic Variants | % 5 Kb Upstream Variants | % 5 Kb Downstream Variants | %Intron Variants | % Genic Variants | Genes with HI Variants | ||
|---|---|---|---|---|---|---|---|---|
| High Impact 1 | Moderate Impact 2 | Low Impact 3 | ||||||
| Vouves bottom | 55.30 | 20.63 | 14.90 | 5.12 | 0.23 | 2.26 | 1.56 | 21,559 |
| Vouves top | 46.53 | 16.81 | 12.98 | 4.15 | 0.19 | 1.87 | 1.31 | 18,618 |
| Kalamon | 59.45 | 18.32 | 13.72 | 4.46 | 0.27 | 2.32 | 1.45 | 25,917 |
| Koroneiki | 59.93 | 18.25 | 13.64 | 4.32 | 0.26 | 2.20 | 1.40 | 26,334 |
| Mastoidis | 60.12 | 18.01 | 13.56 | 4.34 | 0.26 | 2.27 | 1.43 | 25,041 |
| Mavreya | 59.77 | 18.11 | 13.69 | 4.43 | 0.27 | 2.28 | 1.44 | 26,558 |
| Megaritiki | 59.45 | 18.43 | 13.76 | 4.45 | 0.26 | 2.23 | 1.40 | 28,567 |
| Myrtolia | 59.79 | 18.28 | 13.67 | 4.32 | 0.26 | 2.26 | 1.42 | 26,164 |
| Sample | Genes with High Impact Variants | ||||||
|---|---|---|---|---|---|---|---|
| Exon Lost | Frameshift | Splice Acceptor | Splice Donor | Start Lost | Stop Gain | Stop Lost | |
| Vouves Bottom | 2 | 12,651 | 4697 | 4632 | 2112 | 11,624 | 3126 |
| Vouves Top | 1 | 10,588 | 3781 | 3723 | 1792 | 9748 | 2598 |
| Kalamon | 1 | 14,752 | 5954 | 5790 | 2860 | 17,086 | 3603 |
| Koroneiki | 2 | 16,355 | 6503 | 6204 | 3275 | 17,037 | 4003 |
| Mastoidis | 3 | 14,896 | 5798 | 5570 | 2757 | 16,170 | 3593 |
| Mavreya | 5 | 15,829 | 6318 | 6071 | 3111 | 17,307 | 3736 |
| Megaritiki | 3 | 16,924 | 6956 | 6629 | 3386 | 19,322 | 4199 |
| Myrtolia | 1 | 14,565 | 5789 | 5451 | 2747 | 16,332 | 3427 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bombarely, A.; Doulis, A.G.; Lambrou, K.K.; Zioutis, C.; Margaritis, E.; Koubouris, G. Elucidation of the Origin of the Monumental Olive Tree of Vouves in Crete, Greece. Plants 2021, 10, 2374. https://doi.org/10.3390/plants10112374
Bombarely A, Doulis AG, Lambrou KK, Zioutis C, Margaritis E, Koubouris G. Elucidation of the Origin of the Monumental Olive Tree of Vouves in Crete, Greece. Plants. 2021; 10(11):2374. https://doi.org/10.3390/plants10112374
Chicago/Turabian StyleBombarely, Aureliano, Andreas G. Doulis, Katerina K. Lambrou, Christos Zioutis, Evi Margaritis, and Georgios Koubouris. 2021. "Elucidation of the Origin of the Monumental Olive Tree of Vouves in Crete, Greece" Plants 10, no. 11: 2374. https://doi.org/10.3390/plants10112374