Polymer-Nanodiscs as a Novel Alignment Medium for High-Resolution NMR-Based Structural Studies of Nucleic Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of DMPC Liposomes
2.2. SMA-EA Polymer
2.3. Preparation of Polymer-Nanodiscs
2.4. Characterization of SMA-EA Nanodiscs
2.4.1. 1H NMR
2.4.2. Dynamic Light Scattering (DLS)
2.5. NMR Sample Preparation
2.6. RDC NMR Measurements
2.7. Measuring Nanodisc-Induced RDCs in Holo Fluoride Riboswitch
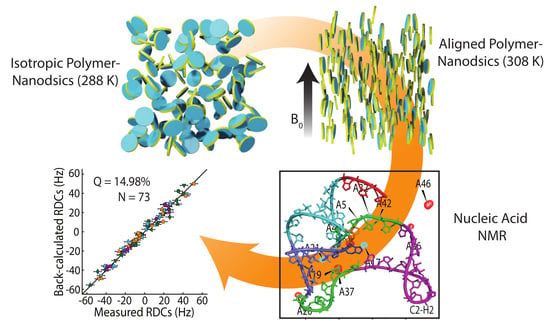
3. Results and Discussion
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prestegard, J.H.; Bougault, C.M.; Kishore, A.I. Residual dipolar couplings in structure determination of biomolecules. Chem. Rev. 2004, 104, 3519–3540. [Google Scholar] [CrossRef]
- Tolman, J.R.; Ruan, K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem. Rev. 2006, 106, 1720–1736. [Google Scholar] [CrossRef]
- Chen, K.; Tjandra, N. The use of residual dipolar coupling in studying proteins by NMR. NMR Proteins Small Biomol. 2012, 326, 47–67. [Google Scholar] [CrossRef] [Green Version]
- Cole, C.A.; Daigham, N.S.; Liu, G.; Montelione, G.T.; Valafar, H. REDCRAFT: A computational platform using residual dipolar coupling NMR data for determining structures of perdeuterated proteins in solution. PLOS Comput. Biol. 2021, 17, e1008060. [Google Scholar] [CrossRef]
- Tjandra, N. Residual dipolar coupling. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 2213–2221. [Google Scholar]
- Bibow, S.; Carneiro, M.G.; Sabo, T.M.; Schwiegk, C.; Becker, S.; Riek, R.; Lee, D. Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings. Protein Sci. 2014, 23, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Bax, A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003, 12, 1–16. [Google Scholar] [CrossRef]
- Sanders, C.R.; Schwonek, J.P. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry 1992, 31, 8898–8905. [Google Scholar] [CrossRef]
- Tjandra, N.; Bax, A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 1997, 278, 1111–1114. [Google Scholar] [CrossRef] [Green Version]
- Ottiger, M.; Bax, A. Characterization of magnetically oriented phospholipid micelles for measurement of dipolar couplings in macromolecules. J. Biomol. NMR 1998, 12, 361–372. [Google Scholar] [CrossRef]
- Canlas, C.G.; Ma, D.; Tang, P.; Xu, Y. Residual dipolar coupling measurements of transmembrane proteins using aligned low-q bicelles and high-resolution magic angle spinning NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 13294–13300. [Google Scholar] [CrossRef]
- Hansen, M.R.; Mueller, L.; Pardi, A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998, 5, 1065–1074. [Google Scholar] [CrossRef]
- Bernadó, P.; Bertoncini, C.W.; Griesinger, C.; Zweckstetter, M.; Blackledge, M. Defining long-range order and local disorder in native α-synuclein using residual dipolar couplings. J. Am. Chem. Soc. 2005, 127, 17968–17969. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Suh, J.Y.; Grishaev, A.; Ghirlando, R.; Takayama, Y.; Clore, G.M. Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J. Am. Chem. Soc. 2010, 132, 13026–13045. [Google Scholar] [CrossRef] [Green Version]
- Marchant, J.; Bax, A.; Summers, M.F. Accurate measurement of residual dipolar couplings in large RNAs by variable flip angle NMR. J. Am. Chem. Soc. 2018, 140, 6978–6983. [Google Scholar] [CrossRef]
- Robertson, A.J.; Courtney, J.M.; Shen, Y.; Ying, J.; Bax, A. Concordance of X-ray and AlphaFold2 models of SARS-CoV-2 main protease with residual dipolar couplings measured in solution. J. Am. Chem. Soc. 2021, 143, 19306–19310. [Google Scholar] [CrossRef]
- Singh, H.; Shukla, M.; Rao, B.J.; Chary, K.V.R. Flagella as a novel alignment medium for the measurement of residual dipolar couplings in proteins. ChemComm 2013, 49, 11403–11405. [Google Scholar] [CrossRef]
- Sager, E.; Tzvetkova, P.; Gossert, A.D.; Piechon, P.; Luy, B. Determination of configuration and conformation of a reserpine derivative with seven stereogenic centers using molecular dynamics with RDC-derived tensorial constraints. Chem.–Eur. J. 2020, 26, 14435–14444. [Google Scholar] [CrossRef]
- Zong, W.; Li, G.-W.; Cao, J.-M.; Lei, X.; Hu, M.-L.; Sun, H.; Griesinger, C.; Tan, R.X. An alignment medium for measuring residual dipolar couplings in pure DMSO: Liquid crystals from graphene oxide grafted with polymer brushes. Angew. Chem. Int. Ed. 2016, 55, 3690–3693. [Google Scholar] [CrossRef] [Green Version]
- Ravula, T.; Ramamoorthy, A. Magnetic alignment of polymer macro-nanodiscs enables residual-dipolar-coupling-based high-resolution structural studies by NMR spectroscopy. Angew. Chem. Int. Ed. 2019, 58, 14925–14928. [Google Scholar] [CrossRef]
- Ravula, T.; Ramamoorthy, A. Measurement of residual dipolar couplings using magnetically aligned and flipped nanodiscs. Langmuir 2022, 38, 244–252. [Google Scholar] [CrossRef]
- Dufourc, E.J. Bicelles and nanodiscs for biophysical chemistry. BBA-Biomembr. 2021, 1863, 183478. [Google Scholar] [CrossRef] [PubMed]
- Günsel, U.; Hagn, F. Lipid nanodiscs for high-resolution NMR studies of membrane proteins. Chem. Rev. 2021, 122, 9395–9421. [Google Scholar] [CrossRef]
- Ramirez, B.E.; Bax, A. Modulation of the alignment tensor of macromolecules dissolved in a dilute liquid crystalline medium. J. Am. Chem. Soc. 1998, 120, 9106–9107. [Google Scholar] [CrossRef]
- Dürr, U.H.; Gildenberg, M.; Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. Chem. Rev. 2012, 112, 6054–6074. [Google Scholar] [CrossRef] [PubMed]
- Lorieau, J.L.; Maltsev, A.S.; Louis, J.M.; Bax, A. Modulating alignment of membrane proteins in liquid-crystalline and oriented gel media by changing the size and charge of phospholipid bicelles. J. Biomol. NMR 2013, 55, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Al-Hashimi, H.M.; Valafar, H.; Terrell, M.; Zartler, E.R.; Eidsness, M.K.; Prestegard, J.H. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J. Magn. Reson. 2000, 143, 402–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolman, J.R.; Al-Hashimi, H.M.; Kay, L.E.; Prestegard, J.H. Structural and dynamic analysis of residual dipolar coupling data for proteins. J. Am. Chem. Soc. 2001, 123, 1416–1424. [Google Scholar] [CrossRef]
- Tian, F.; Al-Hashimi, H.M.; Craighead, J.L.; Prestegard, J.H. Conformational analysis of a flexible oligosaccharide using residual dipolar couplings. J. Am. Chem. Soc. 2001, 123, 485–492. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Hagn, F.; Wagner, G. Structure refinement and membrane positioning of selectively labeled OmpX in phospholipid nanodiscs. J. Biomol. NMR 2015, 61, 249–260. [Google Scholar] [CrossRef]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [Green Version]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.J.; Grigoriu, S.; Huser, S.; Plückthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Nasr, M.L.; Wagner, G. Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat. Protoc. 2018, 13, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Barnaba, C.; Sahoo, B.R.; Ravula, T.; Medina-Meza, I.G.; Im, S.-C.; Anantharamaiah, G.M.; Waskell, L.; Ramamoorthy, A. Cytochrome-P450-induced ordering of microsomal membranes modulates affinity for drugs. Angew. Chem. Int. Ed. 2018, 57, 3391–3395. [Google Scholar] [CrossRef] [PubMed]
- Gentry, K.A.; Anantharamaiah, G.M.; Ramamoorthy, A. Probing protein–protein and protein–substrate interactions in the dynamic membrane-associated ternary complex of cytochromes P450, b5, and reductase. ChemComm 2019, 55, 13422–13425. [Google Scholar] [CrossRef]
- Krishnarjuna, B.; Yamazaki, T.; Anantharamaiah, G.M.; Ramamoorthy, A. Nanodisc reconstitution of flavin mononucleotide binding domain of cytochrome-P450-reductase enables high-resolution NMR probing. ChemComm 2021, 57, 4819–4822. [Google Scholar] [CrossRef]
- Anada, C.; Ikeda, K.; Egawa, A.; Fujiwara, T.; Nakao, H.; Nakano, M. Temperature- and composition-dependent conformational transitions of amphipathic peptide–phospholipid nanodiscs. J. Colloid Interface Sci. 2021, 588, 522–530. [Google Scholar] [CrossRef]
- Anada, C.; Ikeda, K.; Nakao, H.; Nakano, M. Improvement of thermal stability of amphipathic peptide–phospholipid nanodiscs via lateral association of α-helices by disulfide cross-linking. Langmuir 2022, 38, 6977–6983. [Google Scholar] [CrossRef]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.-P.; Dafforn, T.; Overduin, M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef]
- Oluwole, A.O.; Danielczak, B.; Meister, A.; Babalola, J.O.; Vargas, C.; Keller, S. Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer. Angew. Chem. Int. Ed. 2017, 56, 1919–1924. [Google Scholar] [CrossRef]
- Ravula, T.; Ramadugu, S.K.; Di Mauro, G.; Ramamoorthy, A. Bioinspired, size-tunable self-assembly of polymer-lipid bilayer nanodiscs. Angew. Chem. Int. Ed. 2017, 56, 11466–11470. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.C.L.; Tognoloni, C.; Charlton, J.; Bragginton, E.C.; Rothnie, A.J.; Sridhar, P.; Wheatley, M.; Knowles, T.J.; Arnold, T.; Edler, K.J.; et al. An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale 2018, 10, 10609–10619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, K.M.; Harding, B.D.; Sahu, I.D.; Kearns, M.M.; Stowe, R.B.; Dolan, M.T.; Edelmann, R.E.; Dabney-Smith, C.; Page, R.C.; Konkolewicz, D.; et al. Simple derivatization of RAFT-synthesized styrene-maleic anhydride copolymers for lipid disk formulations. Biomacromolecules 2020, 21, 1274–1284. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.Z.; Ramadugu, S.K.; Cox, S.J.; Ramamoorthy, A. Formation of pH-resistant monodispersed polymer-lipid nanodiscs. Angew. Chem. Int. Ed. 2018, 57, 1342–1345. [Google Scholar] [CrossRef]
- Radoicic, J.; Park, S.H.; Opella, S.J. Macrodiscs comprising SMALPs for oriented sample solid-state NMR spectroscopy of membrane proteins. Biophys. J. 2018, 115, 22–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardin, N.Z.; Ravula, T.; Mauro, G.D.; Ramamoorthy, A. Hydrophobic functionalization of polyacrylic acid as a versatile platform for the development of polymer lipid nanodisks. Small 2019, 15, e1804813. [Google Scholar] [CrossRef]
- Ravula, T.; Kim, J.; Lee, D.-K.; Ramamoorthy, A. Magnetic alignment of polymer nanodiscs probed by solid-state NMR spectroscopy. Langmuir 2020, 36, 1258–1265. [Google Scholar] [CrossRef]
- Ravula, T.; Ramamoorthy, A. Synthesis, characterization, and nanodisc formation of non-ionic polymers. Angew. Chem. Int. Ed. 2021, 60, 16885–16888. [Google Scholar] [CrossRef]
- McCalpin, S.D.; Ravula, T.; Ramamoorthy, A. Saponins form nonionic lipid nanodiscs for protein structural studies by Nuclear Magnetic Resonance Spectroscopy. J. Phys. Chem. Lett. 2022, 13, 1705–1712. [Google Scholar] [CrossRef]
- Galiakhmetov, A.R.; Davern, C.M.; Esteves, R.J.A.; Awosanya, E.O.; Guthrie, Q.A.E.; Proulx, C.; Nevzorov, A.A. Aligned peptoid-based macrodiscs for structural studies of membrane proteins by oriented-sample NMR. Biophys. J. 2022, 121, 3263–3270. [Google Scholar] [CrossRef]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012, 335, 233–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Smith, K.D.; Davis, J.H.; Gordon, P.B.; Breaker, R.R.; Strobel, S.A. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 19018–19023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Guffy, S.L.; Williams, B.; Zhang, Q. An excited state underlies gene regulation of a transcriptional riboswitch. Nat. Chem. Biol. 2017, 13, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Fitzkee, N.C.; Bax, A. Facile measurement of ¹H-¹5N residual dipolar couplings in larger perdeuterated proteins. J. Biomol. NMR 2010, 48, 65–70. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.L.; Al-Hashimi, H.M. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: Towards new long-range orientational constraints. J. Magn. Reson. 2006, 179, 299–307. [Google Scholar] [CrossRef]
- Cornilescu, G.; Marquardt, J.L.; Ottiger, M.; Bax, A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 1998, 120, 6836–6837. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.Z.; Bai, J.; Im, S.C.; Waskell, L.; Ramamoorthy, A. Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs. ChemComm 2018, 54, 9615–9618. [Google Scholar] [CrossRef]
- Krishnarjuna, B.; Ravula, T.; Ramamoorthy, A. Detergent-free extraction, reconstitution and characterization of membrane-anchored cytochrome-b5 in native lipids. ChemComm 2020, 56, 6511–6514. [Google Scholar] [CrossRef]
- Campagne, S.; Gervais, V.; Milon, A. Nuclear magnetic resonance analysis of protein–DNA interactions. J. R. Soc. Interface 2011, 8, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Krishnarjuna, B.; Im, S.C.; Ravula, T.; Marte, J.; Auchus, R.J.; Ramamoorthy, A. Non-ionic inulin-based polymer nanodiscs enable functional reconstitution of a redox complex composed of oppositely charged CYP450 and CPR in a lipid bilayer membrane. Anal. Chem. 2022, 94, 11908–11915. [Google Scholar] [CrossRef] [PubMed]
- Krishnarjuna, B.; Ravula, T.; Ramamoorthy, A. Detergent-free isolation of CYP450-reductase’s FMN-binding domain in E. coli lipid-nanodiscs using a charge-free polymer. ChemComm 2022, 58, 4913–4916. [Google Scholar] [CrossRef] [PubMed]
- Fiori, M.C.; Jiang, Y.; Altenberg, G.A.; Liang, H. Polymer-encased nanodiscs with improved buffer compatibility. Sci. Rep. 2017, 7, 7432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.H.; Wu, J.; Yao, Y.; Singh, C.; Tian, Y.; Marassi, F.M.; Opella, S.J. Membrane proteins in magnetically aligned phospholipid polymer discs for solid-state NMR spectroscopy. BBA-Biomembr. 2020, 1862, 183333. [Google Scholar] [CrossRef]
- Baisden, J.T.; Boyer, J.A.; Zhao, B.; Hammond, S.M.; Zhang, Q. Visualizing a protonated RNA state that modulates microRNA-21 maturation. Nat. Chem. Biol. 2021, 17, 80–88. [Google Scholar] [CrossRef]
- Hansen, M.R.; Hanson, P.; Pardi, A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzymol. 2000, 317, 220–240. [Google Scholar] [CrossRef]
- Krishnarjuna, B.; Ramamoorthy, A. Detergent-free isolation of membrane proteins and strategies to study them in a near-native membrane environment. Biomolecules 2022, 12, 1076. [Google Scholar] [CrossRef]
- Kummerlöwe, G.; Luy, B. Residual dipolar couplings for the configurational and conformational analysis of organic molecules. Annu. Rep. NMR Spectrosc. 2009, 68, 193–232. [Google Scholar] [CrossRef]
- Li, G.-W.; Liu, H.; Qiu, F.; Wang, X.-J.; Lei, X.-X. Residual dipolar couplings in structure determination of natural products. Nat. Prod. Bioprospect. 2018, 8, 279–295. [Google Scholar] [CrossRef]
- Nath, N.; d’Auvergne, E.J.; Griesinger, C. Long-range residual dipolar couplings: A tool for determining the configuration of small molecules. Angew. Chem. Int. Ed. 2015, 54, 12706–12710. [Google Scholar] [CrossRef] [PubMed]
- Chiliveri, S.C.; Robertson, A.J.; Torchia, D.A.; Bax, A. Advances in NMR spectroscopy of weakly aligned biomolecular systems. Chem. Rev. 2022, 122, 9307–9330. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnarjuna, B.; Ravula, T.; Faison, E.M.; Tonelli, M.; Zhang, Q.; Ramamoorthy, A. Polymer-Nanodiscs as a Novel Alignment Medium for High-Resolution NMR-Based Structural Studies of Nucleic Acids. Biomolecules 2022, 12, 1628. https://doi.org/10.3390/biom12111628
Krishnarjuna B, Ravula T, Faison EM, Tonelli M, Zhang Q, Ramamoorthy A. Polymer-Nanodiscs as a Novel Alignment Medium for High-Resolution NMR-Based Structural Studies of Nucleic Acids. Biomolecules. 2022; 12(11):1628. https://doi.org/10.3390/biom12111628
Chicago/Turabian StyleKrishnarjuna, Bankala, Thirupathi Ravula, Edgar M. Faison, Marco Tonelli, Qi Zhang, and Ayyalusamy Ramamoorthy. 2022. "Polymer-Nanodiscs as a Novel Alignment Medium for High-Resolution NMR-Based Structural Studies of Nucleic Acids" Biomolecules 12, no. 11: 1628. https://doi.org/10.3390/biom12111628