Evaluation of the Aquatic Toxicity of Several Triazole Fungicides
Abstract
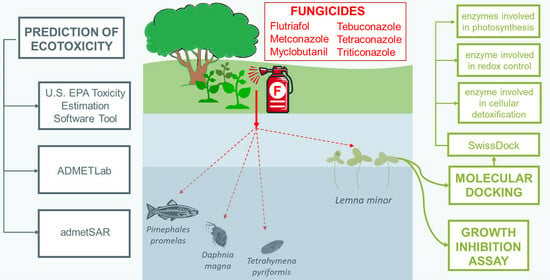
:1. Introduction
2. Materials and Methods
2.1. Materials Used in the Experimental Approach
2.2. Lemna minor Growth Inhibition Assay
2.3. Statistical Analysis Used in the Experimental Approach
2.4. Predictions of the Toxicological Effects of Triazole Fungicides on Aqueous Organisms
2.5. Molecular Docking Study
3. Results and Discussions
3.1. Effects of Triazole Fungicides on Lemna minor
3.2. Prediction of Toxicity of Triazole Fungicides on Other Aqueous Organisms
3.3. Evaluation of the Interactions of Investigated Fungicides with Enzymes Involved in the Photosynthesis Systems, Redox Control and Cellular Detoxification of Lemna minor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pesticide in Global Market Overview 2023–2027. Available online: https://www.reportlinker.com/ (accessed on 25 January 2024).
- Hüesker, F.; Lepenies, R. Why does pesticide pollution in water persist? Environ. Sci. Policy 2022, 128, 185–193. [Google Scholar] [CrossRef]
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An overlooked pesticide class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [CrossRef] [PubMed]
- Fungicides Market Report Analysis & Report Summary. Available online: https://www.marketsandmarkets.com/Market-Reports/fungicides-356.html (accessed on 25 January 2024).
- Del Puerto, O.; Gonçalves, N.P.F.; Medana, C.; Prevot, A.B.; Roslev, P. Attenuation of toxicity and occurrence of degradation products of the fungicide tebuconazole after combined vacuum UV and UVC treatment of drinking water. Environ. Sci. Pollut. Res. 2022, 29, 58312–58325. [Google Scholar] [CrossRef] [PubMed]
- Roman, D.L.; Voiculescu, D.I.; Filip, M.; Ostafe, V.; Isvoran, A. Effects of triazole fungicides on soil microbiota and on the activities of enzymes found in soil: A review. Agriculture 2021, 11, 893. [Google Scholar] [CrossRef]
- Filimon, M.N.; Voia, S.O.; Vladoiu, D.L.; Isvoran, A.; Ostafe, V. Temperature dependent effect of difenoconazole on enzymatic activity from the soil. J. Serb. Chem. Soc. 2015, 80, 1127–1137. [Google Scholar] [CrossRef]
- Westlund, P.; Nasuhoglu, D.; Isazadeh, S.; Yargeau, V. Investigation of acute and chronic toxicity trends of pesticides using high-throughput bioluminescence assay based on the test organism Vibrio fischeri. Arch. Environ. Contam. Toxicol. 2018, 74, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Roman, D.L.; Voiculescu, D.I.; Ostafe, V.; Ciorsac, A.; Isvoran, A. A review of the toxicity of triazole fungicides approved to be used in European Union to the soil and aqueous environment. Ovidius Univ. Ann. Chem. 2022, 33, 113–120. [Google Scholar] [CrossRef]
- Roman, D.L.; Voiculescu, D.I.; Matica, M.A.; Baerle, V.; Filimon, M.N.; Ostafe, V.; Isvoran, A. Assessment of the effects of triticonazole on soil and human health. Molecules 2022, 27, 6554. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, J.; Li, F.; Liu, J. Triazole fungicide tebuconazole disrupts human placental trophoblast cell functions. J. Hazard. Mater. 2016, 308, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Gridan, I.M.; Ciorsac, A.A.; Isvoran, A. Prediction of ADME-Tox properties and toxicological endpoints of triazole fungicides used for cereals protection. ADMET DMPK 2019, 7, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Voiculescu, D.I.; Roman, D.L.; Ostafe, V.; Isvoran, A. A cheminformatics study regarding the human health risks assessment of the stereoisomers of difenoconazole. Molecules 2022, 27, 4682. [Google Scholar] [CrossRef] [PubMed]
- Petit, A.-N.; Fontaine, F.; Vatsa, P.; Clément, C.; Vaillant-Gaveau, N. Fungicide impacts on photosynthesis in crop plants. Photosynth. Res. 2012, 111, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Singh Sidhu, G.P.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K.; et al. Photosynthetic response of plants under different abiotic stresses: A review. J. Plant Groeth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Gorshkov, A.P.; Kusakin, P.G.; Borisov, Y.G.; Tsyganova, A.V.; Tsyganov, V.E. Effect of triazole fungicides titul duo and vintage on the development of pea (Pisum sativum L.) symbiotic nodules. Int. J. Mol. Sci. 2023, 24, 8646. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, P.; Li, J. Enantioselective effects of the fungicide metconazole on photosynthetic activity in Microcystis flos-aquae. Ecotox Environ. Saf. 2021, 211, 111894. [Google Scholar] [CrossRef] [PubMed]
- Shaki, F.; Rezayian, M.; Ebrahimzadeh Maboud, H.; Niknam, V. Role of triazolic compounds in underlying mechanisms of plant stress tolerance; a review. Iran. J. Plant Physiol. 2022, 12, 3943–3954. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Tang, Q.; Mei, L.; Cao, J.; Huang, H.; Zhang, Z. Aquatic ecological risk evaluation of chiral triazole fungicide prothioconazole and its metabolite prothioconazole-desthio on Lemna minor. Sustainability 2022, 14, 16292. [Google Scholar] [CrossRef]
- Alkimin, G.D.D.; Santos, J.; Soares, A.M.V.M.; Nunes, B. Ecotoxicological effects of the azole antifungal agent clotrimazole on the macrophyte species Lemna minor and Lemna gibba. Comb. Biochem. Phys. C 2020, 237, 108835. [Google Scholar] [CrossRef] [PubMed]
- Speck-Planche, A.; Kleandrova, V.V.; Luan, F.; Cordeiro, M.N.D.S. Predicting multiple ecotoxicological profiles in agrochemical fungicides: A multi-species chemoinformatic approach. Ecotox Environ. Saf. 2012, 80, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.M.; Roy, K.; Benfenati, E. Chemometric modeling of Daphnia magna toxicity of agrochemicals. Chemosphere 2019, 224, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Organisation for Economic Co-Operation and Development. Test No. 221: Lemna sp. Growth Inhibition Test. In OECD Guidelines for the Testing of Chemicals 2; OECD: Paris, France, 2006. [Google Scholar]
- Boros, B.-V.; Dascalu, D.; Ostafe, V.; Isvoran, A. Assessment of the effects of chitosan, chitooligosaccharides and their derivatives on Lemna minor. Molecules 2022, 27, 6123. [Google Scholar] [CrossRef] [PubMed]
- Boros, B.-V.; Grau, N.I.; Isvoran, A.; Datcu, A.D.; Ianovici, N.; Ostafe, V. A study of the effects of sodium alginate and sodium carboxymethyl cellulose on the growth of common duckweed (Lemna minor L.): Scientific paper. J. Serb. Chem. Soc. 2022, 87, 657–667. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Technical Overview of Ecological Risk Assessment—Analysis Phase: Ecological Effects Characterization. Available online: https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/technical-overview-ecological-risk-assessment-0 (accessed on 18 January 2024).
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An international database for pesticide risk assessments and management. Hum. Ecol. Risk Assess. Int. J. 2016, 22, 1050–1064. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- United States Environmental Protection Agency. User’s Guide for T. E. S. T. (Toxicity Estimation Software Tool) Version 5.1—A Java Application to Estimate Toxicities and Physical Properties from Molecular Structure. Available online: https://www.epa.gov/sites/default/files/2016-05/documents/600r16058.pdf (accessed on 15 January 2024).
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2018, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res 2022, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Giménez–Moolhuyzen, M.; van der Blom, J.; Lorenzo–Mínguez, P.; Cabello, T.; Crisol–Martínez, E. Photosynthesis inhibiting effects of pesticides on sweet pepper leaves. Insects 2020, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- McCarty, R.E.; Evron, Y.; Johnson, E.A. The chloroplast ATP synthase: A rotary enzyme? Annu. Rev. Plant Phys. 2000, 51, 83–109. [Google Scholar] [CrossRef] [PubMed]
- Fromme, P.; Jordan, P.; Krauß, N. Structure of photosystem I. Biochim. Biophys. Acta 2001, 1507, 5–31. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, P.R. Photosystem I: Function and physiology. Annu. Rev. Plant Phys. 2001, 52, 593–626. [Google Scholar] [CrossRef]
- Azarin, K.; Usatov, A.; Makarenko, M.; Kozel, N.; Kovalevich, A.; Dremuk, I.; Yemelyanova, A.; Logacheva, M.; Fedorenko, A.; Averina, N. A point mutation in the photosystem I P700 chlorophyll a apoprotein A1 gene confers variegation in Helianthus annuus L. Plant Mol. Biol. 2020, 103, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Barber, J. Photosystem II: The engine of life. Q. Rev. Biophys. 2003, 36, 71–89. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, J.P.; Brudvig, G.W. Water-splitting chemistry of photosystem II. Chem. Rev. 2006, 106, 4455–4483. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A. The Calvin cycle revisited. Photosynth. Res. 2003, 75, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef] [PubMed]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; Flutriafol Report; University of Hertfordshire: Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/353.htm (accessed on 15 January 2024).
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; Metconazole Report; University of Hertfordshire: Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/451.htm#2 (accessed on 15 January 2024).
- Marinho, M.D.C.; Diogo, B.S.; Lage, O.M.; Antunes, S.C. Ecotoxicological evaluation of fungicides used in viticulture in non-target organisms. Environ. Sci. Pollut. Res. 2020, 27, 43958–43969. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Huang, L.; Diao, J.; Zhou, Z. Enantioselective toxic effects and degradation of myclobutanil enantiomers in Scenedesmus obliquus. Chirality 2013, 25, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.-G.; Tu, X.; Liu, L.; Wang, G.-X.; Ling, F. The oxidative stress response of myclobutanil and cyproconazole on Tetrahymena thermophila. Environ. Toxicol. Pharmacol. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; Myclobutanil Report; University of Hertfordshire: Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/478.htm#2 (accessed on 15 January 2024).
- Montague, B.; Al-Mudallal, A. Ecological Risk Assessment for Section 3 Registration of Tebuconazole on Wheat, Cucurbits, Bananas, Turnips, Tree Nuts, Hops, and Sunflowers. Available online: https://www3.epa.gov/pesticides/chem_search/cleared_reviews/csr_PC-128997_25-Jul-00_a.pdf (accessed on 11 January 2024).
- European Food Safety Authority. Conclusion on the peer review of the pesticide risk assessment of the active substance tebuconazole. EFSA J. 2014, 12, 3485. [Google Scholar] [CrossRef]
- Tofan, L.; Niță, V.; Nenciu, M.; Coatu, V.; Lazăr, L.; Damir, N.; Vasile, D.; Popoviciu, D.R.; Brotea, A.-G.; Curtean-Bănăduc, A.M.; et al. Multiple assays on non-target organisms to determine the risk of acute environmental toxicity in tebuconazole-based fungicides widely used in the black sea coastal area. Toxics 2023, 11, 597. [Google Scholar] [CrossRef]
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; University of Hertfordshire; Tebuconazole Report; Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/610.htm#2 (accessed on 15 January 2024).
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; Tetraconazole Report; University of Hertfordshire: Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/626.htm#2 (accessed on 15 January 2024).
- Ochoa-Acuña, H.G.; Bialkowski, W.; Yale, G.; Hahn, L. Toxicity of soybean rust fungicides to freshwater algae and Daphnia magna. Ecotoxicology 2009, 18, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Deng, Y.; Zhang, W.; He, R.; Fan, J.; Zhu, W.; Zhou, Z.; Diao, J. Risk assessment of the chiral fungicide triticonazole: Enantioselective effects, toxicity, and fate. J. Agric. Food Chem. 2022, 70, 2712–2721. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Deng, Y.; Zhang, W.; Zhang, L.; Wang, Z.; Li, B.; Diao, J.; Zhou, Z. Enantioselective mechanism of toxic effects of triticonazole against Chlorella pyrenoidosa. Ecotox Environ. Saf. 2019, 185, 109691. [Google Scholar] [CrossRef] [PubMed]
- Agriculture & Environment Research Unit (AERU)—University of Hertfordshire. Pesticide Properties Database; Triticonazole Report; University of Hertfordshire: Hatfield, UK; Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/673.htm#2 (accessed on 15 January 2024).
- Coors, A.; Vollmar, P.; Sacher, F.; Thoma, A. Joint Effects of Pharmaceuticals and Chemicals Regulated under REACH in Wastewater Treatment Plant Effluents—Evaluating Concepts for a Risk Assessment by Means of Experimental Scenarios. Available online: https://www.bmuv.de/fileadmin/Daten_BMU/Pools/Forschungsdatenbank/fkz_3712_64_419_klaeranlagenablaeufe_bf.pdf (accessed on 11 January 2024).
- Roman, D.L.; Matica, M.A.; Ciorsac, A.; Boros, B.V.; Isvoran, A. The effects of the fungicide myclobutanil on soil enzyme activity. Agriculture 2023, 13, 1956. [Google Scholar] [CrossRef]







| Fungicide | Commercial Name | Producer and Country | Tested Concentrations |
|---|---|---|---|
| Flutriafol | Impact | Cheminova A/S, Harboøre, Denmark | 0.02, 0.2, 2, 20 and 200 mg/L |
| Metconazole | Caramba | BASF Agro BV Arnhem, Zürich, Switzerland | 0.002, 0.02, 0.2, 2 and 20 mg/L |
| Myclobutanil | Systhane forte | DowAgroSciences LLC, Indianapolis, IL, USA | 0.03, 0.3, 3, 30 and 300 mg/L |
| Tebuconazole | Sextan | Ascenza Agro S.A., Setúbal, Portugal | 0.003, 0.03, 0.3, 3 and 30 mg/L |
| Tetraconazole | Domark | ISAGRO S.p.A, Milan, Italy | 0.005, 0.5, 0.5, 5 and 50 mg/L |
| Triticonazole | Premis | BASF Agro BV Arnhem, Zürich, Switzerland | 0.1, 1, 10, 100 and 1000 mg/L |
| Enzyme Belonging to Lemna minor, Its Uniprot and AlphaFold IDs | Corresponding Enzyme Having a Determined Structure in Protein Data Bank, Its PDB and Uniprot IDs | RMSD Values for the Superposition of the AlphaFold Model and Experimental Structure |
|---|---|---|
| Enzymes Involved in Photosynthesis | ||
| Chloroplast ATP synthase subunit alpha (A9L981/AF-A9L981-F1) | Chloroplast ATP synthase subunit alpha from Spinacia oleracea in complex with ATP (6VMD chain C/P06450) | 1.305 Å for 388 CA pruned atom pairs from all 436 atom pairs |
| Chloroplast ATP synthase subunit beta (A9L9A3/AF-A9L9A3-F1) | Chloroplast ATP synthase subunit beta from Spinacia oleracea in complex with ATP (6VMD chain D/P00825) | 1.043 Å for 403 CA pruned atom pairs from all 479 atom pairs |
| Photosystem I P700 chlorophyll a apoproteins A1 (A9L996/AF-A9L996-F1) and A2 (A9L995/AF-A9L995-F1) | Photosystem I P700 chlorophyll a apoprotein A1 from Pisum sativum bound in the photosystem I complex and containing beta-carotene, chlorophyll a, and phylloquinone molecules (2WSE chain A/P05310) | 0.898 Å for 609 CA pruned atom pairs from all 730 atom pairs |
| Photosystem II CP43 reaction center protein (A9L992/AF-A9L992-F1) | Photosystem II CP43 reaction center protein from Pisum sativum bound in the photosystem II complex and containing chlorophyll a and beta-carotene molecules (6YP7 chain C/P06004) | 1.031 Å for 440 CA pruned atom pairs from all 450 atom pairs |
| Photosystem II CP47 reaction center protein (A9L9C2/AF-A9L9C2-F1) | Photosystem II CP47 reaction center protein from Pisum sativum bound in the photosystem II complex containing chlorophyll a and beta-carotene molecules (6YP7 chain B/Q9XQR6) | 1.113 Å for 485 CA pruned atom pairs from all 503 atom pairs |
| Photosystem II proteins D1 (A9L976/AF-A9L976-F1) and D2 (A9L991/AF-A9L991-F1) | Photosystem II protein D1 from Pisum sativum bound in the photosystem II complex containing chlorophyll a and beta-carotene molecules (6YP7 chain A/P06585) | 1.070 Å for 320 CA pruned atom pairs from all 334 atom pairs |
| Ribulose bisphosphate carboxylase large chain (A9L9A4/AF-A9L9A4-F1) | Ribulose bisphosphate carboxylase large chain from Spinacia oleracea in complex with ribulose-1,5-diphosphate (1RCX chain B/P00875) | 0.306 Å for 464 CA pruned atom pairs from all 467 atom pairs |
| Enzyme Involved in Redox Control | ||
| Glutathione peroxidase (A5Z284/AF-A5Z284-F1) | Glutathione peroxidase from Schistosoma mansoni in complex with pyrophosphate (2WGR chain A/Q00277) | 0.796 Å for 92 CA pruned atom pairs from all 95 atom pairs |
| Enzyme Involved in Cellular Detoxification | ||
| Glutathione transferase (A0A0F6PRM5/AF-A0A0F6PRM5-F1) | Glutathione transferase from Alopecurus myosuroides in complex with S-hydroxy-glutathione and succinic acid (6RIV chain A/Q9ZS17) | 0.690 Å for 202 pruned atom pairs from all 213 atom pairs |
| Organism/Fungicide | FLU | MET | MYC | TEB | TET | TRI |
|---|---|---|---|---|---|---|
| TEST | ||||||
| Fathead minnow LC50 96 h −log10 (mg/L) | 4.82 | 4.94 | 4.78 | 4.88 | 5.69 | 5.57 |
| Daphnia magna LC50 48 h −log10 (mg/L) | 4.44 | 4.51 | 5.10 | 4.50 | 4.63 | 4.61 |
| ADMETLab2.0 | ||||||
| Fathead minnow LC50 96 h −log10 [(mg/L)/(1000 × MW)] | 3.70 | 4.35 | 4.15 | 3.71 | 5.30 | 3.73 |
| Daphnia magna LC50 48 h −log10 [(mg/L)/(1000 × MW)] | 4.44 | 3.89 | 3.51 | 3.42 | 4.64 | 3.49 |
| Tetrahymena pyriformis IGC50 48 h −log10 [ (mg/L)/(1000 × MW)] | 2.56 | 3.79 | 3.11 | 3.41 | 4.34 | 2.96 |
| admetSAR2.0 | ||||||
| Probability to produce 96 h toxicity against fathead minnow | −0.44 | 0.93 | 0.97 | 0.69 | 0.85 | 0.98 |
| Probability to produce crustacea 48 h aquatic toxicity | 0.61 | −0.50 | 0.69 | 0.66 | 0.61 | 0.55 |
| Tetrahymena pyriformis 48 h −log10 IGC50 (µg/L) | 0.40 | 0.91 | 1.65 | 1.38 | 0.59 | 1.02 |
| Fungicide/Enzyme | Ligand | ΔG (kcal/mol) | |||||
|---|---|---|---|---|---|---|---|
| FLU | MET | MYC | TEB | TET | TRI | ||
| Chloroplast ATP synthase subunit alpha | ATP | - | - | - | - | - | - |
| Chloroplast ATP synthase subunit beta | ADP | −6.25 | −6.21 | −6.10 | −6.77 | −6.49 | −6.81 |
| Photosystem I P700 chlorophyll a apoproteins A1 and A2 | chlorophyll | −7.24 | −6.68 | −6.42 | −6.50 | −7.04 | −7.08 |
| phylloquinone a | - | - | - | - | - | - | |
| Photosystem II proteins D1 and D2 | chlorophyll a | −6.66 | −7.17 | −6.17 | −7.03 | −6.99 | −7.23 |
| β-carotene | - | - | - | - | - | - | |
| pheophytin a | - | - | - | - | - | - | |
| Photosystem II C43 reaction center protein | chlorophyll a | −7.91 | −7.61 | −7.99 | −7.48 | −7.44 | −7.66 |
| β-carotene | - | - | - | - | - | - | |
| Photosystem II C47 reaction center protein | chlorophyll a | −7.65 | −7.47 | −7.62 | −7.44 | −8.28 | −7.60 |
| β-carotene | - | - | - | - | - | - | |
| Ribulose bisphosphate carboxylase large chain | ribulose-1,5-diphosphate | - | - | - | - | - | - |
| Glutathione peroxidase | pyrophosphate 2 | - | - | - | - | - | - |
| Glutathione S-transferase | S-hydroxy-glutathione | −7.38 | −7.40 | −7.38 | −7.99 | −7.60 | −8.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boros, B.-V.; Roman, D.-L.; Isvoran, A. Evaluation of the Aquatic Toxicity of Several Triazole Fungicides. Metabolites 2024, 14, 197. https://doi.org/10.3390/metabo14040197
Boros B-V, Roman D-L, Isvoran A. Evaluation of the Aquatic Toxicity of Several Triazole Fungicides. Metabolites. 2024; 14(4):197. https://doi.org/10.3390/metabo14040197
Chicago/Turabian StyleBoros, Bianca-Vanesa, Diana-Larisa Roman, and Adriana Isvoran. 2024. "Evaluation of the Aquatic Toxicity of Several Triazole Fungicides" Metabolites 14, no. 4: 197. https://doi.org/10.3390/metabo14040197