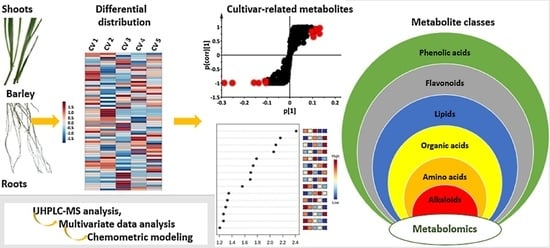
A Metabolomics Approach and Chemometric Tools for Differentiation of Barley Cultivars and Biomarker Discovery
Abstract
:1. Introduction
2. Results
2.1. UHPLC-MS Analyses of Barley Leaf and Root Extracts
2.2. Multivariate Data Analyses: Principal Component- and Hierarchical Clustering Analyses (PCA and HiCA)
2.3. Unravelling the Cultivar-Specific Metabolic Profile of Barley Leaves and Roots
2.3.1. Distribution of Metabolite Classes in Leaf and Root Extracts of Barley Cultivars
2.3.2. Partial Least Squares- and Orthogonal Partial Least Squares-Discriminant Analyses (O)PLS-DA: Differential Metabolite Profiles and Potential Biomarkers
3. Discussion
4. Materials and Methods
4.1. Barley Plant Material and Growth Conditions
4.2. Metabolite Extraction and Pre-Analytical Sample Preparation
4.3. Sample Analysis Using Ultra-High Performance Liquid Chromatography—High Definition Mass Spectrometry
4.4. Data Processing and Data Mining
4.5. Metabolite Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breseghello, F.; Coelho, A.S.G. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J. Agric. Food Chem. 2013, 61, 8277–8286. [Google Scholar] [CrossRef]
- Gienapp, P.; Laine, V.N.; Mateman, A.C.; van Oers, K.; Visser, M.E. Environment-dependent genotype-phenotype associations in avian breeding time. Front. Genet. 2017, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslam, M.M.; Karanja, J.K. Genotype by environment interactions modulate sugarcane response to mechanical wounding stress. Physiol. Mol. Plant Path. 2020, 109, 101443. [Google Scholar] [CrossRef]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, G.G.; Martino-Catt, S.; Glenn, K.C. Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 2007, 3, 259–272. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Diola, V.; Menezes Daloso de, D.; Antunes, W.C. Metabolomics. In Omics in Plant Breeding; Borém, A., Fritsche-Neto, R., Eds.; Wiley: New York, NY, USA, 2014; pp. 1–11. [Google Scholar]
- Deda, O.; Gika, H.; Raikos, N.; Theodoridis, G. GC-MS-based metabolic phenotyping. In The Handbook of Metabolic Phenotyping; Lindon, J.C., Nicholson, J.K., Holmes, E., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 137–169. [Google Scholar]
- Lawas, L.M.F.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice cultivars with different tolerance to combined drought and heat stress under field conditions. Gigascience 2019, 8, giz050. [Google Scholar] [CrossRef]
- Mareya, C.R.; Tugizimana, F.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019, 9, 1840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zeng, X.; Xu, Q.; Mei, X.; Yuan, H.; Jiabu, D.; Sang, Z.; Nyima, T. Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 2019, 11, 21. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, Z.; Li, Y.; Li, Z.; Liu, H.; Zhou, W. Metabolite profiling of sorghum seeds of different colors from different sweet sorghum cultivars using a widely targeted metabolomics approach. Int. J. Genom. 2020, 2020, 6247429. [Google Scholar] [CrossRef]
- Pretorius, C.J.; Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomics for biomarker discovery: Key signatory metabolic profiles for the identification and discrimination of oat cultivars. Metabolites 2021, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Dictionary of Natural Products. Available online: https://dnp.chemnetbase.com (accessed on 31 May 2021).
- Hamany Djande, C.Y.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Habituated Moringa oleifera callus retains metabolic responsiveness to external plant growth regulators. Plant Cell Tissue Organ. Cult. 2019, 137, 249–264. [Google Scholar] [CrossRef]
- Ramabulana, A.T.; Steenkamp, P.; Madala, N.; Dubery, I.A. Profiling of chlorogenic acids from Bidens pilosa and differentiation of closely related positional isomers with the aid of UHPLC-QTOF-MS/MS-based in-source collision-induced dissociation. Metabolites 2020, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N.; Kelly, L.J.; Susan, K.; Nikolai, K. Hierarchical scheme for LC-MSn identification of chlorogenic acids. J. Agric. Food Chem. 2003, 51, 2900–2911. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Sawikowska, A.; Krajewski, P.; Kachlicki, P. Combined mass spectrometric and chromatographic methods for in-depth analysis of phenolic secondary metabolites in barley leaves. J. Mass Spectrom. 2015, 50, 513–532. [Google Scholar] [CrossRef]
- Chemspider—Search and Share Chemistry. Available online: www.chemspider.com (accessed on 31 May 2021).
- Gorzolka, K.; Bednarz, H.; Niehaus, K. Detection and localization of novel hordatine-like compounds and glycosylated derivates of hordatines by imaging mass spectrometry of barley seeds. Planta 2014, 239, 1321–1335. [Google Scholar] [CrossRef]
- Pihlava, J.M.; Kurtelius, T.; Hurme, T. Total hordatine content in different types of beers. J. Inst. Brew. 2016, 122, 212–217. [Google Scholar] [CrossRef]
- PubChem. Available online: http://pubchem.ncbi.nlm.nih.gov (accessed on 31 May 2021).
- Zhang, P.; Chan, W.; Ang, I.L.; Wei, R.; Lam, M.M.; Lei, K.M.; Poon, T.C. Revisiting fragmentation reactions of protonated α-amino acids by high-resolution electrospray ionization tandem mass spectrometry with collision-induced dissociation. Sci. Rep. 2019, 9, 6453. [Google Scholar] [CrossRef]
- Al Kadhi, O.; Melchini, A.; Mithen, R.; Saha, S. Development of a LC-MS/MS method for the simultaneous detection of tricarboxylic acid cycle intermediates in a range of biological matrices. J. Anal. Methods. Chem. 2017, 2017, 5391832. [Google Scholar] [CrossRef] [Green Version]
- MassBank of North America. Available online: https://mona.fiehnlab.ucdavis.edu/spectra/search (accessed on 31 May 2021).
- Enomoto, H.; Sensu, T.; Sato, K.; Sato, F.; Paxton, T.; Yumoto, E.; Miyamoto, K.; Asahina, M.; Yokota, T.; Yamane, H. Visualisation of abscisic acid and 12-oxo-phytodienoic acid in immature Phaseolus vulgaris L. seeds using desorption electrospray ionisation-imaging mass spectrometry. Sci. Rep. 2017, 7, 42977. [Google Scholar] [CrossRef]
- Oliw, E.H.; Hamberg, M. Biosynthesis of jasmonates from linoleic acid by the fungus Fusarium oxysporum. Evidence for a novel allene oxide cyclase. Lipids 2019, 54, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Tasche, D. Validation of internal rating systems and PD estimates. In The Analytics of Risk Model Validation; Christodoulakis, G., Satchell, S., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 169–196. [Google Scholar]
- Westerhuis, J.A.; Hoefsloot, H.C.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.; van Duijnhoven, J.P.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Lee, J.H.; Kim, M.H.; Lee, J.A.; Kim, Y.B.; Jung, E.; Kim, Y.S.; Lee, J.; Park, D. Hordenine, a single compound produced during barley germination, inhibits melanogenesis in human melanocytes. Food Chem. 2013, 141, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Chen, W.; Wang, W.; Zhang, H.; Liu, X.; Luo, J. Comprehensive profiling and natural variation of flavonoids in rice. J. Integr. Plant Biol. 2014, 56, 876–886. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Differential synthesis of secondary metabolites by Streptomyces chrestomyceticus strain ADP4 in response to modulation in nitrogen source and its anti-Candida activity. Proceedings 2020, 66, 5. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Jia, M.; Kang, X.; Wang, S.; Sun, H.; Liu, M.; Wang, A.; Strappe, P.; Zhou, Z. Association of enriched metabolites profile with the corresponding volatile characteristics induced by rice yellowing process. Food Chem. 2021, 349, 129173. [Google Scholar] [CrossRef]
- Dong, X.; Gao, Y.; Chen, W.; Wang, W.; Gong, L.; Liu, X.; Luo, J. Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol. Plant 2015, 8, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Ullah, N.; Yüce, M.; Gökçe, Z.N.Ö.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genom. 2017, 18, 969. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic profiling of the host response of tomato (Solanum lycopersicum) following infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. [Google Scholar] [CrossRef] [Green Version]
- Linatoc, A.C.; Idris, A.; Bakar, M.F.A. Influence of light intensity on the photosynthesis and phenolic contents of Mangifera indica. J. Sci. Technol. 2018, 10, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, B.; Murugan, R.; Parama, V.R.; Vohland, M. Use of different chemometric approaches for an estimation of soil properties at field scale with near infrared spectroscopy. J. Plant Nutr. Soil Sci. 2018, 181, 704–713. [Google Scholar] [CrossRef]
- Büchele, D.; Chao, M.; Ostermann, M.; Leenen, M.; Bald, I. Multivariate chemometrics as a key tool for prediction of K and Fe in a diverse German agricultural soil-set using EDXRF. Sci. Rep. 2019, 9, 17588. [Google Scholar] [CrossRef]
- Xing, J.; Saeys, W.; De Baerdemaeker, J. Combination of chemometric tools and image processing for bruise detection on apples. Comput. Electron. Agric. 2007, 56, 1–13. [Google Scholar] [CrossRef]
- Zarei, I.; Luna, E.; Leach, J.E.; McClung, A.; Vilchez, S.; Koita, O.; Ryan, E.P. Comparative rice bran metabolomics across diverse cultivars and functional rice gene–bran metabolite relationships. Metabolites 2018, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuberger, A.L.; Broeckling, C.D.; Kirkpatrick, K.R.; Prenni, J.E. Application of nontargeted metabolite profiling to discover novel markers of quality traits in an advanced population of malting barley. Plant Biotechnol. J. 2014, 12, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, C.; Monnier, Y.; Santonja, M.; Gallet, C.; Weston, L.A.; Prévosto, B.; Saunier, A.; Baldy, V.; Bousquet-Mélou, A. The impact of competition and allelopathy on the trade-off between plant defense and growth in two contrasting tree species. Front. Plant Sci. 2016, 7, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, B.P.; Meints, B.M.; Hayes, P.M. Organic barley producers’ desired qualities for crop improvement. Org. Agric. 2020, 13, 1–8. [Google Scholar] [CrossRef]
- Sumbele, S.; Fotelli, M.N.; Nikolopoulos, D.; Tooulakou, G.; Liakoura, V.; Liakopoulos, G.; Bresta, P.; Dotsika, E.; Adams, M.A.; Karabourniotis, G. Photosynthetic capacity is negatively correlated with the concentration of leaf phenolic compounds across a range of different species. AoB Plants 2012, 2012, pls025. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, R.A.; Spencer, E.Y.; Unwin, C.H. An antifungal factor from barley of possible significance in disease resistance. Can. J. Bot. 1960, 38, 21–29. [Google Scholar] [CrossRef]
- Stoessl, A.; Unwin, C.H. The antifungal factors in barley. V. Antifungal activity of the hordatines. Can. J. Bot. 1970, 48, 465–470. [Google Scholar] [CrossRef]
- Oueslati, O.; Ben-Hammouda, M.; Ghorbal, M.H.; El Gazzeh, M.; Kremer, R.J. Role of phenolic acids in expression of barley (Hordeum vulgare) autotoxicity. Allelopath. J. 2009, 23, 157–166. [Google Scholar]
- Oueslati, O.; Ben-Hammouda, M.A. Laboratory approach to investigate auto-toxicity of soils cultivated with barley. Allelopath. J. Interact. 2017, 3, 9–14. [Google Scholar]
- Bouhaouel, I.; Richard, G.; Fauconnier, M.L.; Ongena, M.; Franzil, L.; Gfeller, A.; Slim Amara, H.; du Jardin, P. Identification of barley (Hordeum vulgare L. subsp. vulgare) root exudates allelochemicals, their autoallelopathic activity and against Bromus diandrus Roth. Germination. Agronomy 2019, 9, 345. [Google Scholar] [CrossRef] [Green Version]
- Vyvyan, J.R. Allelochemicals as leads for new herbicides and agrochemicals. Tetrahedron 2002, 58, 1631–1646. [Google Scholar] [CrossRef]
- Bertholdsson, N.O. Early vigour and allelopathy—Two useful traits for enhanced barley and wheat competitiveness against weeds. Weed Res. 2005, 45, 94–102. [Google Scholar] [CrossRef]
- Lebecque, S.; Crowet, J.M.; Lins, L.; Delory, B.M.; du Jardin, P.; Fauconnier, M.L.; Deleu, M. Interaction between the barley allelochemical compounds gramine and hordenine and artificial lipid bilayers mimicking the plant plasma membrane. Sci. Rep. 2018, 8, 9784. [Google Scholar] [CrossRef] [Green Version]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [Green Version]
- Davies, K.M.; Jibran, R.; Zhou, Y.; Albert, N.W.; Brummell, D.A.; Jordan, B.R.; Bowman, J.L.; Schwinn, K.E. The evolution of flavonoid biosynthesis: A bryophyte perspective. Front. Plant Sci. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Rupasinghe, T.; Callahan, D.L.; Natera, S.H.; Smith, P.; Hill, C.B.; Roessner, U.; Boughton, B.A. Spatio-temporal metabolite and elemental profiling of salt stressed barley seeds during initial stages of germination by MALDI-MSI and µ-XRF spectrometry. Front. Plant Sci. 2019, 10, 1139. [Google Scholar] [CrossRef] [Green Version]
- Balmer, D.; Flors, V.; Glauser, G.; Mauch-Mani, B. Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front. Plant Sci. 2013, 4, 82. [Google Scholar] [CrossRef] [Green Version]
- Maver, M.; Miras-Moreno, B.; Lucini, L.; Trevisan, M.; Pii, Y.; Cesco, S.; Mimmo, T. New insights in the allelopathic traits of different barley genotypes: Middle Eastern and Tibetan wild-relative accessions vs. cultivated modern barley. PLoS ONE 2020, 15, e0231976. [Google Scholar] [CrossRef]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tayeh, C.; Randoux, B.; Laruelle, F.; Bourdon, N.; Reignault, P. Lipids as markers of induced resistance in wheat: A biochemical and molecular approach. Commun. Agric. Appl. Biol. Sci. 2013, 78, 459–465. [Google Scholar]
- Lopez-Bucio, J.; Nieto-Jacobo, M.F.; Ramırez-Rodrıguez, V.; Herrera-Estrella, L. Organic acid metabolism in plants: From adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 2000, 160, 1–13. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Eprintsev, A.T. Organic acids: The pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci. 2016, 7, 1042. [Google Scholar] [CrossRef] [Green Version]
- Meyer, S.; De Angeli, A.; Fernie, A.R.; Martinoia, E. Intra-and extra-cellular excretion of carboxylates. Trends Plant Sci. 2010, 15, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Joshi, V.; Joung, J.G.; Fei, Z.; Jander, G. Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef]
- Obata, T.; Witt, S.; Lisec, J.; Palacios-Rojas, N.; Florez-Sarasa, I.; Yousfi, S.; Araus, J.L.; Cairns, J.E.; Fernie, A.R. Metabolite profiles of maize leaves in drought, heat and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2015, 169, 2665–2683. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Oh, D.G.; Singh, D.; Lee, J.S.; Lee, S.; Lee, C.H. Exploring the metabolomic diversity of plant species across spatial (leaf and stem) components and phylogenic groups. BMC Plant Biol. 2020, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tohge, T.; Niida, R.; Saito, K. Identification of a flavonol 7-o-rhamnosyltransferase gene determining flavonoid pattern in Aabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 2007, 282, 14932–14941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alawiye, T.T.; Babalola, O.O. Metabolomics: Current application and prospects in crop production. Biologia 2021, 76, 227–239. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Castillo, S.; Gopalacharyulu, P.; Yetukuri, L.; Orešič, M. Algorithms and tools for the preprocessing of LC–MS metabolomics data. Chemom. Intell. Lab. Syst. 2011, 108, 23–32. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- MetaboAnalyst—Statistical, Functional and Integrative Analysis of Metabolomics Data. Available online: www.metaboanalyst.ca (accessed on 31 May 2021).
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Maydeu-Olivares, A.; Garcia-Forero, C. Goodness-of-fit testing. In International Encyclopedia of Education, 3rd ed.; Tierney, R., Peterson, P., Baker, E., McGaw, B., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2010; Volume 7, pp. 190–196. [Google Scholar]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for OPLS models and its applicability in multivariate time series analysis. Chemom. Intell. Lab. Syst. 2015, 146, 297–304. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Berger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamany Djande, C.Y.; Piater, L.A.; Steenkamp, P.A.; Tugizimana, F.; Dubery, I.A. A Metabolomics Approach and Chemometric Tools for Differentiation of Barley Cultivars and Biomarker Discovery. Metabolites 2021, 11, 578. https://doi.org/10.3390/metabo11090578
Hamany Djande CY, Piater LA, Steenkamp PA, Tugizimana F, Dubery IA. A Metabolomics Approach and Chemometric Tools for Differentiation of Barley Cultivars and Biomarker Discovery. Metabolites. 2021; 11(9):578. https://doi.org/10.3390/metabo11090578
Chicago/Turabian StyleHamany Djande, Claude Y., Lizelle A. Piater, Paul A. Steenkamp, Fidele Tugizimana, and Ian A. Dubery. 2021. "A Metabolomics Approach and Chemometric Tools for Differentiation of Barley Cultivars and Biomarker Discovery" Metabolites 11, no. 9: 578. https://doi.org/10.3390/metabo11090578