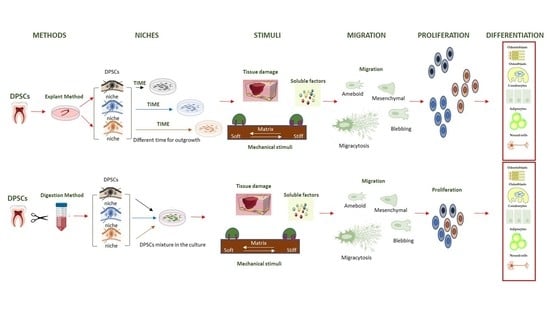
The Migration and the Fate of Dental Pulp Stem Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. HDPSCs
3. Migration
3.1. Lamellipodia
3.2. Filopodia
3.3. Stress Fibers
3.4. Blebbing
3.5. Migrasomes
4. Mechanosensing and Mechanotransduction
5. The hDPSC Fate
6. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitsiadis, T.A.; Catón, J.; Pagella, P.; Orsini, G.; Jimenez-Rojo, L. Monitoring Notch Signaling-Associated Activation of Stem Cell Niches within Injured Dental Pulp. Front. Physiol. 2017, 8, 372. [Google Scholar] [CrossRef] [PubMed]
- Lizier, N.F.; Kerkis, A.; Gomes, C.M.; Hebling, J.; Oliveira, C.F.; Caplan, A.I.; Kerkis, I. Scaling-up of Dental Pulp Stem Cells Isolated from Multiple Niches. PLoS ONE 2012, 7, e39885. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Xin, C.; Zhang, Y.; Yan, J.; Chen, Z.; Xu, H.; Liang, M.; Wu, B.; Fang, F.; Qiu, W. Characterization of Odontogenic Differentiation from Human Dental Pulp Stem Cells Using TMT-Based Proteomic Analysis. BioMed Res. Int. 2020, 2020, 3871496. [Google Scholar] [CrossRef]
- Fujii, Y.; Kawase-Koga, Y.; Hojo, H.; Yano, F.; Sato, M.; Chung, U.-I.; Ohba, S.; Chikazu, D. Bone Regeneration by Human Dental Pulp Stem Cells Using a Helioxanthin Derivative and Cell-Sheet Technology. Stem Cell Res. Ther. 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Li, H.; Wang, R.; Yang, D.; Li, B.; Cao, X.; Fu, J. Transplanted Dental Pulp Stem Cells Migrate to Injured Area and Express Neural Markers in a Rat Model of Cerebral Ischemia. Cell. Physiol. Biochem. 2018, 45, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, H.; Marushima, A.; Ishikawa, H.; Toyomura, J.; Ohyama, A.; Watanabe, M.; Takaoka, S.; Bukawa, H.; Matsumura, A.; Matsumaru, Y.; et al. Induced Neural Cells from Human Dental Pulp Ameliorate Functional Recovery in a Murine Model of Cerebral Infarction. Stem Cell Rev. Rep. 2022, 18, 595–608. [Google Scholar] [CrossRef]
- Driesen, R.B.; Hilkens, P.; Smisdom, N.; Vangansewinkel, T.; Dillen, Y.; Ratajczak, J.; Wolfs, E.; Gervois, P.; Ameloot, M.; Bronckaers, A.; et al. Dental Tissue and Stem Cells Revisited: New Insights From the Expression of Fibroblast Activation Protein-Alpha. Front. Cell Dev. Biol. 2020, 7, 389. [Google Scholar] [CrossRef]
- Nito, C.; Sowa, K.; Nakajima, M.; Sakamoto, Y.; Suda, S.; Nishiyama, Y.; Nakamura-Takahashi, A.; Nitahara-Kasahara, Y.; Ueda, M.; Okada, T.; et al. Transplantation of Human Dental Pulp Stem Cells Ameliorates Brain Damage Following Acute Cerebral Ischemia. Biomed. Pharmacother. 2018, 108, 1005–1014. [Google Scholar] [CrossRef]
- García-Sánchez, D.; Fernández, D.; Rodríguez-Rey, J.C.; Pérez-Campo, F.M. Enhancing Survival, Engraftment, and Osteogenic Potential of Mesenchymal Stem Cells. World J. Stem Cells 2019, 11, 748–763. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal Human Dental Pulp Stem Cells (DPSCs) in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef]
- Al-Maswary, A.A.; O’Reilly, M.; Holmes, A.P.; Walmsley, A.D.; Cooper, P.R.; Scheven, B.A. Exploring the Neurogenic Differentiation of Human Dental Pulp Stem Cells. PLoS ONE 2022, 17, e0277134. [Google Scholar] [CrossRef] [PubMed]
- Mousawi, F.; Peng, H.; Li, J.; Ponnambalam, S.; Roger, S.; Zhao, H.; Yang, X.; Jiang, L.H. Chemical Activation of the Piezo1 Channel Drives Mesenchymal Stem Cell Migration via Inducing ATP Release and Activation of P2 Receptor Purinergic Signaling. Stem Cells 2020, 38, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Montoya, C.; Orrego, S.; Wei, X.; Ling, J.; Lelkes, P.I.; Yang, M. Topographic Cues of a Novel Bilayered Scaffold Modulate Dental Pulp Stem Cells Differentiation by Regulating YAP Signalling through Cytoskeleton Adjustments. Cell Prolif. 2019, 52, e12676. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, M.; Kimura, M.; Ouchi, T.; Nakamura, T.; Ohyama, S.; Ando, M.; Nomura, S.; Azuma, T.; Ichinohe, T.; Shibukawa, Y. Mechanical Stimulation-Induced Calcium Signaling by Piezo1 Channel Activation in Human Odontoblast Reduces Dentin Mineralization. Front. Physiol. 2021, 12, 704518. [Google Scholar] [CrossRef] [PubMed]
- Di Scipio, F.; Sprio, A.E.; Folino, A.; Carere, M.E.; Salamone, P.; Yang, Z.; Berrone, M.; Prat, M.; Losano, G.; Rastaldo, R.; et al. Injured Cardiomyocytes Promote Dental Pulp Mesenchymal Stem Cell Homing. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2152–2161. [Google Scholar] [CrossRef]
- Martens, W.; Wolfs, E.; Struys, T.; Politis, C.; Bronckaers, A.; Lambrichts, I. Expression Pattern of Basal Markers in Human Dental Pulp Stem Cells and Tissue. Cells Tissues Organs 2012, 196, 490–500. [Google Scholar] [CrossRef]
- Li, K.; O’Dwyer, R.; Yang, F.; Cymerman, J.; Li, J.; Feldman, J.D.; Simon, M.; Rafailovich, M. Enhancement of Acellular Biomineralization, Dental Pulp Stem Cell Migration, and Differentiation by Hybrid Fibrin Gelatin Scaffolds. Dent. Mater. 2023, 39, 305–319. [Google Scholar] [CrossRef]
- Küçükkaya Eren, S.; Bahador Zirh, E.; Zirh, S.; Sharafi, P.; Zeybek, N.D. Combined Effects of Bone Morphogenetic Protein-7 and Mineral Trioxide Aggregate on the Proliferation, Migration, and Differentiation of Human Dental Pulp Stem Cells. J. Appl. Oral Sci. 2022, 30, e20220086. [Google Scholar] [CrossRef]
- Gronthos, S.; Arthur, A.; Bartold, P.M.; Shi, S. A Method to Isolate and Culture Expand Human Dental Pulp Stem Cells. In Mesenchymal Stem Cell Assays and Applications; Vemuri, M., Chase, L., Rao, M., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 107–121. ISBN 978-1-60761-999-4. [Google Scholar]
- Patil, V.R.; Kharat, A.H.; Kulkarni, D.G.; Kheur, S.M.; Bhonde, R.R. Long Term Explant Culture for Harvesting Homogeneous Population of Human Dental Pulp Stem Cells. Cell Biol. Int. 2018, 42, 1602–1610. [Google Scholar] [CrossRef]
- Lott, K.; Collier, P.; Ringor, M.; Howard, K.M.; Kingsley, K. Administration of Epidermal Growth Factor (EGF) and Basic Fibroblast Growth Factor (BFGF) to Induce Neural Differentiation of Dental Pulp Stem Cells (DPSC) Isolates. Biomedicines 2023, 11, 255. [Google Scholar] [CrossRef]
- Xiao, N.; Yu, W.Y.; Liu, D. Glial Cell-Derived Neurotrophic Factor Promotes Dental Pulp Stem Cell Migration. J. Tissue Eng. Regen. Med. 2018, 12, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Bonnamain, V.; Thinard, R.; Sergent-Tanguy, S.; Huet, P.; Bienvenu, G.; Naveilhan, P.; Farges, J.C.; Alliot-Licht, B. Human Dental Pulp Stem Cells Cultured in Serum-Free Supplemented Medium. Front. Physiol. 2013, 4, 357. [Google Scholar] [CrossRef]
- Hilkens, P.; Gervois, P.; Fanton, Y.; Vanormelingen, J.; Martens, W.; Struys, T.; Politis, C.; Lambrichts, I.; Bronckaers, A. Effect of Isolation Methodology on Stem Cell Properties and Multilineage Differentiation Potential of Human Dental Pulp Stem Cells. Cell Tissue Res. 2013, 353, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, J.-Y.; Park, H.-J.; Kim, M.-K.; Kim, Y.-I.; Kim, H.-J.; Bae, S.-K.; Bae, M.-K. Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2019, 20, 5778. [Google Scholar] [CrossRef]
- Fatima, N.; Khan, A.A.; Vishwakarma, S.K. Immunophenotypic and Molecular Analysis of Human Dental Pulp Stem Cells Potential for Neurogenic Differentiation. Contemp. Clin. Dent. 2017, 8, 81–89. [Google Scholar] [CrossRef]
- Huang, G.T.J.; Sonoyama, W.; Chen, J.; Park, S.H. In Vitro Characterization of Human Dental Pulp Cells: Various Isolation Methods and Culturing Environments. Cell Tissue Res. 2006, 324, 225–236. [Google Scholar] [CrossRef]
- Senthilkumar, S.; Venugopal, C.; Parveen, S.; Shobha, K.; Rai, K.S.; Kutty, B.M.; Dhanushkodi, A. Remarkable Migration Propensity of Dental Pulp Stem Cells towards Neurodegenerative Milieu: An in Vitro Analysis. Neurotoxicology 2020, 81, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Lew, W.Z.; Feng, S.W.; Lin, C.T.; Huang, H.M. Use of 0.4-Tesla Static Magnetic Field to Promote Reparative Dentine Formation of Dental Pulp Stem Cells through Activation of P38 MAPK Signalling Pathway. Int. Endod. J. 2019, 52, 28–43. [Google Scholar] [CrossRef]
- Alliot-Licht, B.; Hurtrel, D.; Gregoire, M. Characterization of A-Smooth Muscle Actin Positive Cells in Mineralized Human Dental Pulp Cultures. Arch. Oral Biol. 2001, 46, 221–228. [Google Scholar] [CrossRef]
- Pagella, P.; Nombela-arrieta, C.; Mitsiadis, T.A. Distinct Expression Patterns of Cxcl12 in Mesenchymal Stem Cell Niches of Intact and Injured Rodent Teeth. Int. J. Mol. Sci. 2021, 22, 3024. [Google Scholar] [CrossRef]
- Wei, X.-L.; Luo, L.; Chen, M.-Z.; Zhou, J.; Lan, B.-Y.; Ma, X.-M.; Chen, W.-X. Temporospatial Expression of Neuropeptide Substance P in Dental Pulp Stem Cells During Odontoblastic Differentiation In Vitro and Reparative Dentinogenesis In Vivo. J. Endod. 2023, 49, 276–285. [Google Scholar] [CrossRef]
- Ehlinger, C.; Mathieu, E.; Rabineau, M.; Ball, V.; Lavalle, P.; Haikel, Y.; Vautier, D.; Kocgozlu, L. Insensitivity of Dental Pulp Stem Cells Migration to Substrate Stiffness. Biomaterials 2021, 275, 120969. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.; Seol, D.; Gomez-Contreras, P.C.; Keen, H.L.; Shin, K.; Martin, J.A. Exosome-Based Cell Homing and Angiogenic Differentiation for Dental Pulp Regeneration. Int. J. Mol. Sci. 2023, 24, 466. [Google Scholar] [CrossRef] [PubMed]
- Rustom, A. The Missing Link: Does Tunnelling Nanotube-Based Supercellularity Provide a New Understanding of Chronic and Lifestyle Diseases? Open Biol. 2016, 6, 160057. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tian, Z.; Liu, Q.; Wang, T.; Ban, L.K.; Lee, H.H.C.; Umezawa, A.; Almansour, A.I.; Arumugam, N.; Kumar, R.S.; et al. Neuronal Cell Differentiation of Human Dental Pulp Stem Cells on Synthetic Polymeric Surfaces Coated with ECM Proteins. Front. Cell Dev. Biol. 2022, 10, 893241. [Google Scholar] [CrossRef]
- Zhang, S.; Ye, D.; Ma, L.; Ren, Y.; Dirksen, R.T.; Liu, X. Purinergic Signaling Modulates Survival/Proliferation of Human Dental Pulp Stem Cells. J. Dent. Res. 2019, 98, 242–249. [Google Scholar] [CrossRef]
- Miyazaki, A.; Sugimoto, A.; Yoshizaki, K.; Kawarabayashi, K.; Iwata, K.; Kurogoushi, R.; Kitamura, T.; Otsuka, K.; Hasegawa, T.; Akazawa, Y.; et al. Coordination of WNT Signaling and Ciliogenesis during Odontogenesis by Piezo Type Mechanosensitive Ion Channel Component 1. Sci. Rep. 2019, 9, 14762. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, L.; Chen, L.; Jiang, J.; Zhou, X.; Wang, M.; Fan, Y. Static Magnetic Field Regulates Proliferation, Migration, Differentiation, and YAP/TAZ Activation of Human Dental Pulp Stem Cells. J. Tissue Eng. Regen. Med. 2018, 12, 2029–2040. [Google Scholar] [CrossRef]
- Yang, J.-W.; Zhang, Y.-F.; Wan, C.-Y.; Sun, Z.-Y.; Nie, S.; Jian, S.-J.; Zhang, L.; Song, G.-T.; Chen, Z. Autophagy in SDF-1α-Mediated DPSC Migration and Pulp Regeneration. Biomaterials 2015, 44, 11–23. [Google Scholar] [CrossRef]
- Du, L.; Feng, R.; Ge, S. PTH/SDF-1α Cotherapy Promotes Proliferation, Migration and Osteogenic Differentiation of Human Periodontal Ligament Stem Cells. Cell Prolif. 2016, 49, 599–608. [Google Scholar] [CrossRef]
- Liang, C.; Liang, Q.; Xu, X.; Liu, X.; Gao, X.; Li, M.; Yang, J.; Xing, X.; Huang, H.; Tang, Q.; et al. Bone Morphogenetic Protein 7 Mediates Stem Cells Migration and Angiogenesis: Therapeutic Potential for Endogenous Pulp Regeneration. Int. J. Oral Sci. 2022, 14, 38. [Google Scholar] [CrossRef]
- Li, J.; Diao, S.; Yang, H.; Cao, Y.; Du, J.; Yang, D. IGFBP5 Promotes Angiogenic and Neurogenic Differentiation Potential of Dental Pulp Stem Cells. Dev. Growth Differ. 2019, 61, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.; Yoon, J.Y.; Park, J.H.; Lee, H.H.; Dashnyam, K.; Kim, H.W.; Lee, J.H.; Shin, J.S.; Kim, J. Bin The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study. Cells 2022, 11, 3398. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Duan, M.; Guo, D.; Du, X.; Zhang, D.; Xie, J. Microenvironmental Stiffness Mediates Cytoskeleton Re-Organization in Chondrocytes through Laminin-FAK Mechanotransduction. Int. J. Oral Sci. 2022, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liang, A.; Zhou, Y.; Huang, Y.; Liao, C.; Zhang, X.; Gong, Q. Hypoxia Preconditioned DPSC-Derived Exosomes Regulate Angiogenesis via Transferring LOXL2. Exp. Cell Res. 2023, 425, 113543. [Google Scholar] [CrossRef]
- Wang, D.; Lyu, Y.; Yang, Y.; Zhang, S.; Chen, G.; Pan, J.; Tian, W. Schwann Cell-Derived EVs Facilitate Dental Pulp Regeneration through Endogenous Stem Cell Recruitment via SDF-1/CXCR4 Axis. Acta Biomater. 2022, 140, 610–624. [Google Scholar] [CrossRef]
- Laudani, S.; La Cognata, V.; Iemmolo, R.; Bonaventura, G.; Villaggio, G.; Saccone, S.; Barcellona, M.L.; Cavallaro, S.; Sinatra, F. Effect of a Bone Marrow-Derived Extracellular Matrix on Cell Adhesion and Neural Induction of Dental Pulp Stem Cells. Front. Cell Dev. Biol. 2020, 8, 100. [Google Scholar] [CrossRef]
- Hendijani, F. Explant Culture: An Advantageous Method for Isolation of Mesenchymal Stem Cells from Human Tissues. Cell Prolif. 2017, 50, e12334. [Google Scholar]
- Kok, Z.Y.; Alaidaroos, N.Y.A.; Alraies, A.; Colombo, J.S.; Davies, L.C.; Waddington, R.J.; Sloan, A.J.; Moseley, R. Dental Pulp Stem Cell Heterogeneity: Finding Superior Quality “Needles” in a Dental Pulpal “Haystack” for Regenerative Medicine-Based Applications. Stem Cells Int. 2022, 2022, 9127074. [Google Scholar] [CrossRef]
- Marrelli, M.; Codispoti, B.; Shelton, R.M.; Scheven, B.A.; Cooper, P.R.; Tatullo, M.; Paduano, F. Dental Pulp Stem Cell Mechanoresponsiveness: Effects of Mechanical Stimuli on Dental Pulp Stem Cell Behavior. Front. Physiol. 2018, 9, 1685. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Ren, J.Y.; Yang, B. Priming Strategies for Controlling Stem Cell Fate: Applications and Challenges in Dental Tissue Regeneration. World J. Stem Cells 2021, 13, 1625–1646. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, M.; Thurakkal, B.; Spatz, J.P.; Das, T. Dynamic Heterogeneity Influences the Leader-Follower Dynamics during Epithelial Wound Closure: Heterogeneity Begets Better Coordination. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190391. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Hense, B.A.; Fuchs, T.M.; Utz, M.; Pötzsche, C. Bet-Hedging in Stochastically Switching Environments. J. Theor. Biol. 2013, 336, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Yang, D.; Yi, W.; Cao, H.; Xiao, G. Roles of Leader and Follower Cells in Collective Cell Migration. Mol. Biol. Cell 2021, 32, 1267–1272. [Google Scholar] [CrossRef]
- Callan-Jones, A. Self-Organization in Amoeboid Motility. Front. Cell Dev. Biol. 2022, 10, 1000071. [Google Scholar] [CrossRef] [PubMed]
- Loy, N.; Preziosi, L. Modelling Physical Limits of Migration by a Kinetic Model with Non-Local Sensing. J. Math. Biol. 2020, 80, 1759–1801. [Google Scholar] [CrossRef]
- Loy, N.; Preziosi, L. Kinetic Models with Non-Local Sensing Determining Cell Polarization and Speed According to Independent Cues. J. Math. Biol. 2020, 80, 373–421. [Google Scholar] [CrossRef]
- Conte, M.; Loy, N. Multi-Cue Kinetic Model with Non-Local Sensing for Cell Migration on a Fiber Network with Chemotaxis. Bull. Math. Biol. 2022, 84, 42. [Google Scholar] [CrossRef]
- Ventura, G.; Sedzinski, J. Emerging Concepts on the Mechanical Interplay between Migrating Cells and Microenvironment in Vivo. Front. Cell Dev. Biol. 2022, 10, 1763. [Google Scholar] [CrossRef]
- Rombouts, C.; Jeanneau, C.; Bakopoulou, A.; About, I. Dental Pulp Stem Cell Recruitment Signals within Injured Dental Pulp Tissue. Dent. J. 2016, 4, 8. [Google Scholar] [CrossRef]
- Fortunato, I.C.; Sunyer, R. The Forces behind Directed Cell Migration. Biophysica 2022, 2, 46. [Google Scholar] [CrossRef]
- Vincent, L.G.; Choi, Y.S.; Alonso-Latorre, B.; Del Álamo, J.C.; Engler, A.J. Mesenchymal Stem Cell Durotaxis Depends on Substrate Stiffness Gradient Strength. Biotechnol. J. 2013, 8, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.H.; Choi, O.; Taylor-Weiner, H.; Fuhrmann, A.; Karpiak, J.V.; Almutairi, A.; Engler, A.J. Haptotaxis Is Cell Type Specific and Limited by Substrate Adhesiveness. Cell. Mol. Bioeng. 2015, 8, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Hadden, W.J.; Young, J.L.; Holle, A.W.; McFetridge, M.L.; Kim, D.Y.; Wijesinghe, P.; Taylor-Weiner, H.; Wen, J.H.; Lee, A.R.; Bieback, K.; et al. Stem Cell Migration and Mechanotransduction on Linear Stiffness Gradient Hydrogels. Proc. Natl. Acad. Sci. USA 2017, 114, 5647–5652. [Google Scholar] [CrossRef] [PubMed]
- Espina, J.A.; Marchant, C.L.; Barriga, E.H. Durotaxis: The Mechanical Control of Directed Cell Migration. FEBS J. 2022, 289, 2736–2754. [Google Scholar] [CrossRef]
- Williams, T.D.; Rousseau, A. Actin Dynamics in Protein Homeostasis. Biosci. Rep. 2022, 42, BSR20210848. [Google Scholar] [CrossRef]
- Schaks, M.; Giannone, G.; Rottner, K. Actin Dynamics in Cell Migration. Essays Biochem. 2019, 63, 483–495. [Google Scholar] [CrossRef]
- Chugh, P.; Paluch, E.K. The Actin Cortex at a Glance. J. Cell Sci. 2018, 131, jcs186254. [Google Scholar] [CrossRef]
- Victor Small, J.; Stradal, T.; Vignal, E.; Rottner, K. The lamellipodium: Where motility begins. Trends Cell Biol. 2002, 12, 112–120. [Google Scholar] [CrossRef]
- Rottner, K.; Schaks, M. Assembling Actin Filaments for Protrusion. Curr. Opin. Cell Biol. 2019, 56, 53–63. [Google Scholar] [CrossRef]
- Webb, D.J.; Brown, C.M.; Horwitz, A.F. Illuminating Adhesion Complexes in Migrating Cells: Moving toward a Bright Future. Curr. Opin. Cell Biol. 2003, 15, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Legerstee, K.; Houtsmuller, A.B. A Layered View on Focal Adhesions. Biology 2021, 10, 1189. [Google Scholar] [CrossRef] [PubMed]
- Gallop, J.L. Filopodia and Their Links with Membrane Traffic and Cell Adhesion. Semin. Cell Dev. Biol. 2020, 102, 81–89. [Google Scholar] [CrossRef]
- Houdusse, A.; Titus, M.A. The Many Roles of Myosins in Filopodia, Microvilli and Stereocilia. Curr. Biol. 2021, 31, R586–R602. [Google Scholar] [CrossRef] [PubMed]
- Collart-Dutilleul, P.Y.; Panayotov, I.; Secret, E.; Cunin, F.; Gergely, C.; Cuisinier, F.; Martin, M. Initial Stem Cell Adhesion on Porous Silicon Surface: Molecular Architecture of Actin Cytoskeleton and Filopodial Growth. Nanoscale Res. Lett. 2014, 9, 564. [Google Scholar] [CrossRef]
- Abounit, S.; Zurzolo, C. Wiring through Tunneling Nanotubes—From Electrical Signals to Organelle Transfer. J. Cell Sci. 2012, 125, 1089–1098. [Google Scholar] [CrossRef]
- Sherer, N.M. Long-Distance Relationships: Do Membrane Nanotubes Regulate Cell-Cell Communication and Disease Progression? Mol. Biol. Cell 2013, 24, 1095–1098. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K. Plasticity of Cell Migration: A Multiscale Tuning Model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef]
- Lehtimäki, J.I.; Rajakylä, E.K.; Tojkander, S.; Lappalainen, P. Generation of Stress Fibers through Myosin-Driven Reorganization of the Actin Cortex. Elife 2021, 10, e60710. [Google Scholar] [CrossRef]
- Vallenius, T. Actin Stress Fibre Subtypes in Mesenchymal-Migrating Cells. Open Biol. 2013, 3, 130001. [Google Scholar] [CrossRef]
- Wu, J.; Kent, I.A.; Shekhar, N.; Chancellor, T.J.; Mendonca, A.; Dickinson, R.B.; Lele, T.P. Actomyosin Pulls to Advance the Nucleus in a Migrating Tissue Cell. Biophys. J. 2014, 106, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.G.; Yenepalli, A.; Denais, C.M.; Rape, A.; Beach, J.R.; Wang, Y.-L.; Schiemann, W.P.; Baskaran, H.; Lammerding, J.; Egelhoff, T.T. Non-Muscle Myosin IIB Is Critical for Nuclear Translocation during 3D Invasion. J. Cell Biol. 2015, 210, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin Stress Fibers—Assembly, Dynamics and Biological Roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Livne, A.; Geiger, B. The Inner Workings of Stress Fibers—From Contractile Machinery to Focal Adhesions and Back. J. Cell Sci. 2016, 129, 1293–1304. [Google Scholar] [CrossRef]
- Schick, J.; Raz, E. Blebs—Formation, Regulation, Positioning, and Role in Amoeboid Cell Migration. Front. Cell Dev. Biol. 2022, 10, 926394. [Google Scholar] [CrossRef]
- Srivastava, N.; Traynor, D.; Piel, M.; Kabla, A.J.; Kay, R.R. Pressure Sensing through Piezo Channels Controls Whether Cells Migrate with Blebs or Pseudopods. Proc. Natl. Acad. Sci. USA 2020, 117, 2506–2512. [Google Scholar] [CrossRef]
- Ohgushi, M.; Sasai, Y. Lonely Death Dance of Human Pluripotent Stem Cells: ROCKing between Metastable Cell States. Trends Cell Biol. 2011, 21, 274–282. [Google Scholar] [CrossRef]
- Weng, N.J.H.; Talbot, P. The P2X7 Receptor Is an Upstream Regulator of Dynamic Blebbing and a Pluripotency Marker in Human Embryonic Stem Cells. Stem Cell Res. 2017, 23, 39–49. [Google Scholar] [CrossRef]
- Nicotera, P.; Hartzell, P.; Davis, G.; Orrenius, S. The Formation of Plasma Membrane Blebs in Hepatocytes Exposed to Agents That Increase Cytosolic Ca2+ Is Mediated by the Activation of a Non-Lysosomal Proteolytic System. FEBS Lett. 1986, 209, 139–144. [Google Scholar] [CrossRef]
- Zheng, L.M.; Zychlinsky, A.; Liu, C.C.; Ojcius, D.M.; Young, J.D. Extracellular ATP as a Trigger for Apoptosis or Programmed Cell Death. J. Cell Biol. 1991, 112, 279–288. [Google Scholar] [CrossRef]
- Verhoef, P.A.; Estacion, M.; Schilling, W.; Dubyak, G.R. P2X7 Receptor-Dependent Blebbing and the Activation of Rho-Effector Kinases, Caspases, and IL-1β Release. J. Immunol. 2003, 170, 5728–5738. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the Migrasome, an Organelle Mediating Release of Cytoplasmic Contents during Cell Migration. Cell Res. 2015, 25, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zucker, B.; Zhang, S.; Elias, S.; Zhu, Y.; Chen, H.; Ding, T.; Li, Y.; Sun, Y.; Lou, J.; et al. Migrasome Formation Is Mediated by Assembly of Micron-Scale Tetraspanin Macrodomains. Nat. Cell Biol. 2019, 21, 991–1002. [Google Scholar] [CrossRef]
- Jiang, D.; Jiang, Z.; Lu, D.; Wang, X.; Liang, H.; Zhang, J.; Meng, Y.; Li, Y.; Wu, D.; Huang, Y.; et al. Migrasomes Provide Regional Cues for Organ Morphogenesis during Zebrafish Gastrulation. Nat. Cell Biol. 2019, 21, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yu, L. Migrasome Biogenesis and Functions. FEBS J. 2022, 289, 7246–7254. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zou, Q.; Huang, R.; Li, Y.; Xing, X.; Fang, J.; Ma, L.; Li, L.; Yang, X.; Yu, L. Lateral Transfer of MRNA and Protein by Migrasomes Modifies the Recipient Cells. Cell Res. 2021, 31, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Shi, X.; Zhao, K.; Wang, L.; Shi, K.; Liu, Y.J.; Li, H.; Ji, B.; Jiu, Y. Cell Migration Orchestrates Migrasome Formation by Shaping Retraction Fibers. J. Cell Biol. 2022, 221, e202109168. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Wang, T. How the Mechanical Microenvironment of Stem Cell Growth Affects Their Differentiation: A Review. Stem Cell Res. Ther. 2022, 13, 415. [Google Scholar] [CrossRef]
- Murthy, S.E.; Dubin, A.E.; Patapoutian, A. Piezos Thrive under Pressure: Mechanically Activated Ion Channels in Health and Disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 771–783. [Google Scholar] [CrossRef]
- Xiao, B. Levering Mechanically Activated Piezo Channels for Potential Pharmacological Intervention. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 195–218. [Google Scholar] [CrossRef]
- Wilde, C.; Mitgau, J.; Suchý, T.; Schöneberg, T.; Liebsche, I. Translating the Force-Mechano-Sensing GPCRs Wilde C 2022. Am. J. Physiol. Cell Physiol. 2022, 322, C1047–C1060. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.; Biswas, A.; Soni, G.V. Cellular and Nuclear Forces: An Overview. In Molecular Motors: Methods and Protocols; Lavelle, C., Ed.; Springer: New York, NY, USA, 2018; pp. 1–29. ISBN 978-1-4939-8556-2. [Google Scholar]
- Choi, D.; Park, E.; Jung, E.; Cha, B.; Lee, S.; Yu, J.; Kim, P.M.; Lee, S.; Hong, Y.J.; Koh, C.J.; et al. Piezo1 Incorporates Mechanical Force Signals into the Genetic Program That Governs Lymphatic Valve Development and Maintenance. JCI Insight 2019, 4, e125068. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Mohan, K.; Iglesias, P.A.; Robinson, D.N. Molecular Mechanisms of Cellular Mechanosensing. Nat. Mater. 2013, 12, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T.; et al. Measuring Mechanical Tension across Vinculin Reveals Regulation of Focal Adhesion Dynamics. Nature 2010, 466, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Nourse, J.L.; Pathak, M.M. How Cells Channel Their Stress: Interplay between Piezo1 and the Cytoskeleton. Semin. Cell Dev. Biol. 2017, 71, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ellefsen, K.L.; Holt, J.R.; Chang, A.C.; Nourse, J.L.; Arulmoli, J.; Mekhdjian, A.H.; Abuwarda, H.; Tombola, F.; Flanagan, L.A.; Dunn, A.R.; et al. Myosin-II Mediated Traction Forces Evoke Localized Piezo1-Dependent Ca2+ Flickers. Commun. Biol. 2019, 2, 298. [Google Scholar] [CrossRef]
- Wei, L.; Mousawi, F.; Li, D.; Roger, S.; Li, J.; Yang, X.; Jiang, L.H. Adenosine Triphosphate Release and P2 Receptor Signaling in PiEzO1 Channel-Dependent Mechanoregulation. Front. Pharmacol. 2019, 10, 1304. [Google Scholar] [CrossRef]
- Sun, J.; Ramnath, R.D.; Tamizhselvi, R.; Bhatia, M. Role of Protein Kinase C and Phosphoinositide 3-Kinase-Akt in Substance P-Induced Proinflammatory Pathways in Mouse Macrophages. FASEB J. 2009, 23, 997–1010. [Google Scholar] [CrossRef]
- Peng, H.; Hao, Y.; Mousawi, F.; Roger, S.; Li, J.; Sim, J.A.; Ponnambalam, S.; Yang, X.; Jiang, L.H. Purinergic and Store-Operated Ca2+ Signaling Mechanisms in Mesenchymal Stem Cells and Their Roles in ATP-Induced Stimulation of Cell Migration. Stem Cells 2016, 34, 2102–2114. [Google Scholar] [CrossRef]
- Sugimoto, A.; Miyazaki, A.; Kawarabayashi, K.; Shono, M.; Akazawa, Y.; Hasegawa, T.; Ueda-Yamaguchi, K.; Kitamura, T.; Yoshizaki, K.; Fukumoto, S.; et al. Piezo Type Mechanosensitive Ion Channel Component 1 Functions as a Regulator of the Cell Fate Determination of Mesenchymal Stem Cells. Sci. Rep. 2017, 7, 17696. [Google Scholar] [CrossRef]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef] [PubMed]
- La Noce, M.; Stellavato, A.; Vassallo, V.; Cammarota, M.; Laino, L.; Desiderio, V.; Del Vecchio, V.; Nicoletti, G.F.; Tirino, V.; Papaccio, G.; et al. Hyaluronan-Based Gel Promotes Human Dental Pulp Stem Cells Bone Differentiation by Activating YAP/TAZ Pathway. Cells 2021, 10, 2899. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhang, D.; Zhou, C.; Yuan, Q.; Ye, L.; Zhou, X. Substrate Elasticity Regulates Adipose-Derived Stromal Cell Differentiation towards Osteogenesis and Adipogenesis through β-Catenin Transduction. Acta Biomater. 2018, 79, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Burdick, J.A. Influence of Three-Dimensional Hyaluronic Acid Microenvironments on Mesenchymal Stem Cell Chondrogenesis. Tissue Eng. Part A 2009, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Du, L.; Ge, S. PTH/SDF-1α Cotherapy Induces CD90+CD34− Stromal Cells Migration and Promotes Tissue Regeneration in a Rat Periodontal Defect Model. Sci. Rep. 2016, 6, 30403. [Google Scholar] [CrossRef]
- Hong, H.S.; Kim, S.; Lee, S.; Woo, J.S.; Lee, K.H.; Cheng, X.W.; Son, Y.; Kim, W. Substance-P Prevents Cardiac Ischemia-Reperfusion Injury by Modulating Stem Cell Mobilization and Causing Early Suppression of Injury-Mediated Inflammation. Cell. Physiol. Biochem. 2019, 52, 40–56. [Google Scholar] [CrossRef]
- Nam, D.; Park, A.; Dubon, M.J.; Yu, J.; Kim, W.; Son, Y.; Park, K.S. Coordinated Regulation of Mesenchymal Stem Cell Migration by Various Chemotactic Stimuli. Int. J. Mol. Sci. 2020, 21, 8561. [Google Scholar] [CrossRef]
- Tran, P.B.; Banisadr, G.; Ren, D.; Chenn, A.; Miller, R.J. Chemokine Receptor Expression by Neural Progenitor Cells in Neurogenic Regions of Mouse Brain. J. Comp. Neurol. 2007, 500, 1007–1034. [Google Scholar] [CrossRef]
- Firouzi, N.; Yavari, H.R.; Rahimi, S.; Roshangar, L.; Chitsazha, R.; Amini, M. Concentrated Growth Factors Combined with Lipopolysaccharide Stimulate the In Vitro Regenerative and Osteogenic Activities of Human Dental Pulp Stem Cells by Balancing Inflammation. Int. J. Dent. 2022, 2022, 2316666. [Google Scholar] [CrossRef]


| Digestion Method | Explant Method | Not Specified | Cell Line |
|---|---|---|---|
| [3] | [1] | [8] | [11] |
| [5] | [2] | [12] | [13] |
| [6] | [7] | [14] | [15] |
| [10] | [16] | [17] | [18] |
| [19] | [20] | [21] | [22] |
| [23] | [24] | [25] | |
| [26] | [27] | [28] | |
| [20] | [29] | ||
| [27] | [30] | ||
| [31] | [32] | ||
| [33] | [34] | ||
| [35] | [36] | ||
| [37] | |||
| [38] | |||
| [39] | |||
| [40] | |||
| [41] | |||
| [42] | |||
| [43] | |||
| [44] | |||
| [45] | |||
| [46] | |||
| [47] | |||
| [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lampiasi, N. The Migration and the Fate of Dental Pulp Stem Cells. Biology 2023, 12, 742. https://doi.org/10.3390/biology12050742
Lampiasi N. The Migration and the Fate of Dental Pulp Stem Cells. Biology. 2023; 12(5):742. https://doi.org/10.3390/biology12050742
Chicago/Turabian StyleLampiasi, Nadia. 2023. "The Migration and the Fate of Dental Pulp Stem Cells" Biology 12, no. 5: 742. https://doi.org/10.3390/biology12050742