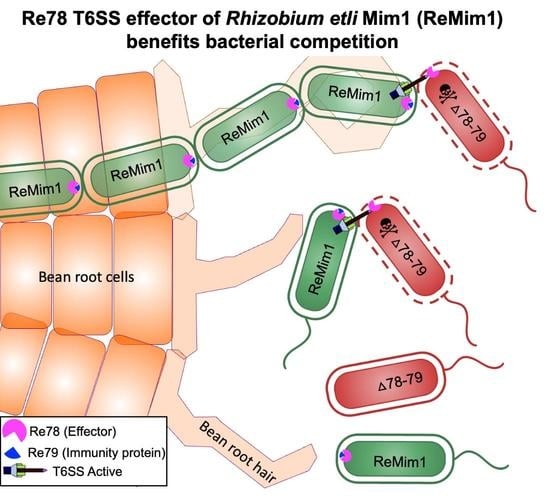
The T6SS-Dependent Effector Re78 of Rhizobium etli Mim1 Benefits Bacterial Competition
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Construction of T6SS Mutants
2.2.1. Generation of Mutants ∆re78–79 and ∆re84–89
2.2.2. Mutant re82HD-AA
2.3. Plasmid Constructions to Express re78 and re79 in E. coli and ReMim1
2.4. E. coli Toxicity Assay
2.5. E. coli Staining for Microscope Viewing
2.6. Subcellular Fractionation and Immunodetection of Re79 Protein from Rhizobium
2.7. Plant Assays
2.8. Bacterial Competition Assay
2.8.1. Competitiveness in Free Living
2.8.2. Competitiveness in Symbiosis
2.9. T6SS Expression of ReMim1
2.9.1. Expression of Promoter Region
2.9.2. RNA Extraction, cDNA Synthesis, and qRT-PCR of T6SS Genes
2.10. Preparation and Analysis of Secretomes
2.11. Bioinformatic Analysis
2.12. Statistical Analysis
3. Results
3.1. Bioinformatic Analysis of Genes That Could Encode Effectors in the Re Mim1 T6SS Gene Cluster
3.2. Symbiotic Phenotype of ReMim1 Mutants in Genes That Could Encode T6SS-Dependent-Effectors
3.3. T6SS Gene Expression
3.4. Proteins Re78/79: A New Toxin/Immunity Pair
3.5. Re78/Re79 Are Involved in Bacterial Competition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From Saprophytes to Endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Deakin, W.J.; Broughton, W.J. Symbiotic Use of Pathogenic Strategies: Rhizobial Protein Secretion Systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef]
- Nelson, M.S.; Sadowsky, M.J. Secretion Systems and Signal Exchange between Nitrogen-Fixing Rhizobia and Legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Lucke, M.; Correa, M.G.; Levy, A. The Role of Secretion Systems, Effectors, and Secondary Metabolites of Beneficial Rhizobacteria in Interactions With Plants and Microbes. Front. Plant Sci. 2020, 11, 589416. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The Rhizobial Type 3 Secretion System: The Dr. Jekyll and Mr. Hyde in the Rhizobium–Legume Symbiosis. Int. J. Mol. Sci. 2022, 23, 11089. [Google Scholar] [CrossRef]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and Regulation of the Type VI Secretion System. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef]
- Coulthurst, S. The Type VI Secretion System: A Versatile Bacterial Weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Singh, R.P.; Kumari, K. Bacterial Type VI Secretion System (T6SS): An Evolved Molecular Weapon with Diverse Functionality. Biotechnol. Lett. 2023, 45, 309–331. [Google Scholar] [CrossRef]
- Schneider, J.P.; Nazarov, S.; Adaixo, R.; Liuzzo, M.; Ringel, P.D.; Stahlberg, H.; Basler, M. Diverse Roles of TssA-like Proteins in the Assembly of Bacterial Type VI Secretion Systems. EMBO J. 2019, 38, e100825. [Google Scholar] [CrossRef]
- Hernandez, R.E.; Gallegos-Monterrosa, R.; Coulthurst, S.J. Type VI Secretion System Effector Proteins: Effective Weapons for Bacterial Competitiveness. Cell Microbiol. 2020, 22, e13241. [Google Scholar] [CrossRef]
- Unni, R.; Pintor, K.L.; Diepold, A.; Unterweger, D. Presence and Absence of Type VI Secretion Systems in Bacteria. Microbiology 2022, 168, 001151. [Google Scholar] [CrossRef]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A Widespread Bacterial Type VI Secretion Effector Superfamily Identified Using a Heuristic Approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Long, M.; Shen, X. Effector–Immunity Pairs Provide the T6SS Nanomachine Its Offensive and Defensive Capabilities. Molecules 2018, 23, 1009. [Google Scholar] [CrossRef]
- Bingle, L.E.; Bailey, C.M.; Pallen, M.J. Type VI Secretion: A Beginner’s Guide. Curr. Opin. Microbiol. 2008, 11, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI Secretion Systems in Plant-Associated Bacteria. Environ. Microbiol. 2018, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, B.F.S.; Castellane, T.C.L.; Tighilt, L.; de Macedo Lemos, E.G.; Rey, L. Rhizobial Exopolysaccharides and Type VI Secretion Systems: A Promising Way to Improve Nitrogen Acquisition by Legumes. Front. Agron. 2021, 3, 661468. [Google Scholar] [CrossRef]
- Gallegos-Monterrosa, R.; Coulthurst, S.J. The Ecological Impact of a Bacterial Weapon: Microbial Interactions and the Type VI Secretion System. FEMS Microbiol. Rev. 2021, 45, fuab033. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-Blocking Genes of a Symbiotic Rhizobium leguminosarum Strain That Are Involved in Temperature-Dependent Protein Secretion. Mol. Plant Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef]
- de Campos, S.B.; Lardi, M.; Gandolfi, A.; Eberl, L.; Pessi, G. Mutations in Two Paraburkholderia phymatum Type VI Secretion Systems Cause Reduced Fitness in Interbacterial Competition. Front. Microbiol. 2017, 8, 2473. [Google Scholar] [CrossRef]
- Lin, H.H.; Huang, H.M.; Yu, M.; Lai, E.M.; Chien, H.L.; Liu, C. Te Functional Exploration of the Bacterial Type VI Secretion System in Mutualism: Azorhizobium caulinodans ORS571– Sesbania Rostrata as a Research Model. Mol. Plant Microbe Interact. 2018, 31, 856–867. [Google Scholar] [CrossRef]
- Salinero-Lanzarote, A.; Pacheco-Moreno, A.; Domingo-Serrano, L.; Durán, D.; Ormeño-Orrillo, E.; Martínez-Romero, E.; Rey, L. The Type VI Secretion System of Rhizobium etli Mim1 Has a Positive Effect in Symbiosis. FEMS Microbiol. Ecol. 2019, 95, fiz054. [Google Scholar] [CrossRef] [PubMed]
- Tighilt, L.; Boulila, F.; De Sousa, B.F.S.; Giraud, E.; Ruiz-Argüeso, T.; Palacios, J.M.; Imperial, J.; Rey, L. The Bradyrhizobium sp. LmicA16 Type VI Secretion System Is Required for Efficient Nodulation of Lupinus spp. Microb. Ecol. 2022, 84, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria Handbook; Blackwell Scientific Publisher: Oxford, UK, 1970. [Google Scholar]
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Wheatley, R.M.; Ramachandran, V.K.; Geddes, B.A.; Perry, B.J.; Yost, C.K.; Poole, P.S. Role of O2 in the Growth of Rhizobium leguminosarum bv. viciae 3841 on Glucose and Succinate. J. Bacteriol. 2017, 199, e00572-16. [Google Scholar] [CrossRef] [PubMed]
- O’Gara, F.; Shanmugam, K.T. Regulation of Nitrogen Fixation by Rhizobia Export of Fixed N2 as NH4+. BBA Gen. Subj. 1976, 437, 313–321. [Google Scholar] [CrossRef]
- Hartig, S.M. Basic Image Analysis and Manipulation in ImageJ. Curr. Protoc. Mol. Biol. 2013, 2, 14.15.1–14.15.12. [Google Scholar] [CrossRef]
- Sibinelli-Sousa, S.; Hespanhol, J.T.; Nicastro, G.G.; Matsuyama, B.Y.; Mesnage, S.; Patel, A.; de Souza, R.F.; Guzzo, C.R.; Bayer-Santos, E. A Family of T6SS Antibacterial Effectors Related to L,D-Transpeptidases Targets the Peptidoglycan. Cell Rep. 2020, 31, 107813. [Google Scholar] [CrossRef]
- Ruiz-Argüeso, T.; Hanus, J.; Evans, H.J. Hydrogen Production and Uptake by Pea Nodules as Affected by Strains of Rhizobium leguminosarum. Arch. Microbiol. 1978, 116, 113–118. [Google Scholar] [CrossRef]
- Miller, J.H. Assay of β-galactosidase. In Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1972. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Moreno, M.L.; Escobar, J.; Izquierdo-Álvarez, A.; Gil, A.; Pérez, S.; Pereda, J.; Zapico, I.; Vento, M.; Sabater, L.; Marina, A.; et al. Disulfide Stress: A Novel Type of Oxidative Stress in Acute Pancreatitis. Free Radic. Biol. Med. 2014, 70, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Pisa, D.; Marina, A.I.; Morato, E.; Rábano, A.; Rodal, I.; Carrasco, L. Evidence for Fungal Infection in Cerebrospinal Fluid and Brain Tissue from Patients with Amyotrophic Lateral Sclerosis. Int. J. Biol. Sci. 2015, 11, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Bondage, D.D.; Lin, J.S.; Ma, L.S.; Kuo, C.H.; Lai, E.M. VgrG C Terminus Confers the Type VI Effector Transport Specificity and Is Required for Binding with PAAR and Adaptor-Effector Complex. Proc. Natl. Acad. Sci. USA 2016, 113, E3931–E3940. [Google Scholar] [CrossRef]
- Pissaridou, P.; Allsopp, L.P.; Wettstadt, S.; Howard, S.A.; Mavridou, D.A.I.; Filloux, A. The Pseudomonas aeruginosa T6SS-VgrG1b Spike Is Topped by a PAAR Protein Eliciting DNA Damage to Bacterial Competitors. Proc. Natl. Acad. Sci. USA 2018, 115, 12519–12524. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.S.; Hachani, A.; Lin, J.S.; Filloux, A.; Lai, E.M. Agrobacterium tumefaciens Deploys a Superfamily of Type VI Secretion DNase Effectors as Weapons for Interbacterial Competition in Planta. Cell Host Microbe 2014, 16, 94–104. [Google Scholar] [CrossRef]
- Zhang, D.; Iyer, L.M.; Aravind, L. A Novel Immunity System for Bacterial Nucleic Acid Degrading Toxins and Its Recruitment in Various Eukaryotic and DNA Viral Systems. Nucleic Acids Res. 2011, 39, 4532–4552. [Google Scholar] [CrossRef]
- Durán, D.; Bernal, P.; Vazquez-Arias, D.; Blanco-Romero, E.; Garrido-Sanz, D.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. Pseudomonas fluorescens F113 Type VI Secretion Systems Mediate Bacterial Killing and Adaption to the Rhizosphere Microbiome. Sci. Rep. 2021, 11, 5772. [Google Scholar] [CrossRef]
- Boak, E.N.; Kirolos, S.; Pan, H.; Pierson, L.S.; Pierson, E.A. The Type VI Secretion Systems in Plant-Beneficial Bacteria Modulate Prokaryotic and Eukaryotic Interactions in the Rhizosphere. Front. Microbiol. 2022, 13, 843092. [Google Scholar] [CrossRef]
- Fei, N.; Ji, W.; Yang, L.; Yu, C.; Qiao, P.; Yan, J.; Guan, W.; Yang, Y.; Zhao, T. Hcp of the Type VI Secretion System (T6SS) in Acidovorax Citrulli Group II Strain Aac5 Has a Dual Role as a Core Structural Protein and an Effector Protein in Colonization, Growth Ability, Competition, Biofilm Formation, and Ferric Iron Absorption. Int. J. Mol. Sci. 2022, 23, 9632. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.B.T. FLS2: An LRR Receptor-like Kinase Involved in the Perception of the Bacterial Elicitor Flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Jana, B.; Salomon, D. Type VI Secretion System: A Modular Toolkit for Bacterial Dominance. Future Microbiol. 2019, 14, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Gao, Z.; Zhang, T.; Zhang, H.; Dong, Y. Crystal Structure of the Type VI Immunity Protein Tdi1 (Atu4351) from Agrobacterium tumefaciens. Acta Crystallogr. F Struct. Biol. Commun. 2019, 75, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, S.; Simmons, E.L.; Verster, A.J.; Mcclure, E.A.; Royce, D.B.; Trus, E.; Swartz, K.; Schultz, D.; Nadell, C.D.; Ross, B.D. Community Composition and the Environment Modulate the Population Dynamics of Type VI Secretion in Human Gut Bacteria. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kirchberger, P.C.; Unterweger, D.; Provenzano, D.; Pukatzki, S.; Boucher, Y. Sequential Displacement of Type VI Secretion System Effector Genes Leads to Evolution of Diverse Immunity Gene Arrays in Vibrio cholerae. Sci. Rep. 2017, 7, 45133. [Google Scholar] [CrossRef] [PubMed]
- Hersch, S.J.; Manera, K.; Dong, T.G. Defending against the Type Six Secretion System: Beyond Immunity Genes. Cell Rep. 2020, 33, 108259. [Google Scholar] [CrossRef]
- Abby, S.S.; Cury, J.; Guglielmini, J.; Néron, B.; Touchon, M.; Rocha, E.P.C. Identification of Protein Secretion Systems in Bacterial Genomes. Sci. Rep. 2016, 6, 23080. [Google Scholar] [CrossRef]
- Lien, Y.-W.; Lai, E.-M. Type VI Secretion Effectors: Methodologies and Biology. Front. Cell. Infect. Microbiol. 2017, 7, 254. [Google Scholar] [CrossRef]
- Lopez, J.; Ly, P.M.; Feldman, M.F. The Tip of the VgrG Spike Is Essential to Functional Type VI Secretion System Assembly in Acinetobacter baumannii. mBio 2020, 11, e02761-19. [Google Scholar] [CrossRef]
- Li, Y.; Yan, X.; Tao, Z. Two Type VI Secretion DNase Effectors Are Utilized for Interbacterial Competition in the Fish Pathogen Pseudomonas plecoglossicida. Front. Microbiol. 2022, 13, 869278. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sun, M.; Pan, Z.; Lu, C.; Yao, H. Diverse Toxic Effectors Are Harbored by VgrG Islands for Interbacterial Antagonism in Type VI Secretion System. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Hood, R.D.; Bui, N.K.; Leroux, M.; Vollmer, W.; Mougous, J.D. Type VI Secretion Delivers Bacteriolytic Effectors to Target Cells. Nature 2011, 475, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, Z.; Du, X.; Shi, Y.; Dang, J.; Lee, M.; Hesek, D.; Mobashery, S.; Wu, M.; Liang, H. A Type VI Secretion System Delivers a Cell Wall Amidase to Target Bacterial Competitors. Mol. Microbiol. 2020, 114, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Dar, Y.; Jana, B.; Bosis, E.; Salomon, D. A Binary Effector Module Secreted by a Type VI Secretion System. EMBO Rep. 2022, 23, e53981. [Google Scholar] [CrossRef]
- Custodio, R.; Ford, R.M.; Ellison, C.J.; Liu, G.; Mickute, G.; Tang, C.M.; Exley, R.M. Type VI Secretion System Killing by Commensal Neisseria Is Influenced by Expression of Type Four Pili. eLife 2021, 10, e63755. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Jia, M.; Liu, J.; Zhou, T.; Wu, L.; Dong, T.; Cai, Z.; Qu, J.; Liu, Y.; Yang, L.; et al. Acquisition of T6SS Effector TseL Contributes to the Emerging of Novel Epidemic Strains of Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 11, e0330822. [Google Scholar] [CrossRef]
- Russell, A.B.; Leroux, M.; Hathazi, K.; Agnello, D.M.; Ishikawa, T.; Wiggins, P.A.; Wai, S.N.; Mougous, J.D. Diverse Type VI Secretion Phospholipases Are Functionally Plastic Antibacterial Effectors. Nature 2013, 496, 508–512. [Google Scholar] [CrossRef]
- Mariano, G.; Trunk, K.; Williams, D.J.; Monlezun, L.; Strahl, H.; Pitt, S.J.; Coulthurst, S.J. A Family of Type VI Secretion System Effector Proteins That Form Ion-Selective Pores. Nat. Commun. 2019, 10, 5484. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Bernal, P.; Allsopp, L.P.; Filloux, A.; Llamas, M.A. The Pseudomonas putida T6SS Is a Plant Warden against Phytopathogens. ISME J. 2017, 11, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.M.; Potthoff, D.B.; Schäfer, M.; Barandun, N.; Vorholt, J.A. Protective Role of the Arabidopsis Leaf Microbiota against a Bacterial Pathogen. Nat. Microbiol. 2021, 6, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Hug, S.; Liu, Y.; Heiniger, B.; Bailly, A.; Ahrens, C.H.; Eberl, L.; Pessi, G. Differential Expression of Paraburkholderia phymatum Type VI Secretion Systems (T6SS) Suggests a Role of T6SS-b in Early Symbiotic Interaction. Front. Plant Sci. 2021, 12, 699590. [Google Scholar] [CrossRef] [PubMed]
- Speare, L.; Cecere, A.G.; Guckes, K.R.; Smith, S.; Wollenberg, M.S.; Mandel, M.J.; Miyashiro, T.; Septer, A.N. Bacterial Symbionts Use a Type VI Secretion System to Eliminate Competitors in Their Natural Host. Proc. Natl. Acad. Sci. USA 2018, 115, E8528–E8537. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hidalgo, P.; Hirsch, A.M. The Nodule Microbiome: N2 fixing Rhizobia Do Not Live Alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Crosbie, D.B.; Mahmoudi, M.; Radl, V.; Brachmann, A.; Schloter, M.; Kemen, E.; Marín, M. Microbiome Profiling Reveals That Pseudomonas Antagonises Parasitic Nodule Colonisation of Cheater Rhizobia in Lotus. New Phytol. 2022, 234, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Lee, H.H.; Park, I.; Seo, Y.S. Genome-Wide Analysis of Type VI System Clusters and Effectors in Burkholderia Species. Plant Pathol. J. 2018, 34, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Z.; Warren, E.A.; Haas, A.L.; Sánchez Peña, A.; Kiedrowski, M.R.; Lomenick, B.; Chou, T.-F.; Bomberger, J.M.; Tirrell, D.A.; Limoli, D.H. Staphylococcal Secreted Cytotoxins Are Competition Sensing Signals for Pseudomonas aeruginosa. bioRxiv 2023. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for in Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Ditta, G.; Stanfield, S.; Corbin, D.; Helinski, D.R.; Donald Helinskl, C.R. Broad Host Range DNA Cloning System for Gram-Negative Bacteria: Construction of a Gene Bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 1980, 77, 7347–7351. [Google Scholar] [CrossRef]
- Wang, E.T.; Rogel, M.A.; García-de los Santos, A.; Martínez-Romero, J.; Cevallos, M.A.; Martínez-Romero, E. Rhizobium etli bv. mimosae, a Novel Biovar Isolated from Mimosa affinis. Int. J. Syst. Bacteriol. 1999, 49, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small Mobilizable Multi-Purpose Cloning Vectors Derived from the Escherichia coli Plasmids PK18 and PK19: Selection of Defined Deletions in the Chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.-M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose P BAD. J. Bacteriol. Res. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Tett, A.J.; Rudder, S.J.; Bourdès, A.; Karunakaran, R.; Poole, P.S. Regulatable Vectors for Environmental Gene Expression in Alphaproteobacteria. Appl. Environ. Microbiol. 2012, 78, 7137–7140. [Google Scholar] [CrossRef]
- Spaink, H.P.; Okker, R.J.H.; Wijffelman, C.A.; Pees, E.; Lugtenberg, B.J.J. Promoters in the Nodulation Region of the Rhizobium leguminosarum Sym Plasmid PRL1JI. Plant Mol. Biol. 1987, 9, 27–39. [Google Scholar] [CrossRef]
- Cheng, H.-P.; Walker, G.C. Succinoglycan Is Required for Initiation and Elongation of Infection Threads during Nodulation of Alfalfa by Rhizobium meliloti. J. Bacteriol. 1998, 180, 5183–5191. [Google Scholar] [CrossRef]
- Gage, D.J. Analysis of Infection Thread Development Using Gfp and DsRed-Expressing Sinorhizobium meliloti. J. Bacteriol. 2002, 184, 7042–7046. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Sousa, B.F.S.; Domingo-Serrano, L.; Salinero-Lanzarote, A.; Palacios, J.M.; Rey, L. The T6SS-Dependent Effector Re78 of Rhizobium etli Mim1 Benefits Bacterial Competition. Biology 2023, 12, 678. https://doi.org/10.3390/biology12050678
De Sousa BFS, Domingo-Serrano L, Salinero-Lanzarote A, Palacios JM, Rey L. The T6SS-Dependent Effector Re78 of Rhizobium etli Mim1 Benefits Bacterial Competition. Biology. 2023; 12(5):678. https://doi.org/10.3390/biology12050678
Chicago/Turabian StyleDe Sousa, Bruna Fernanda Silva, Lucía Domingo-Serrano, Alvaro Salinero-Lanzarote, José Manuel Palacios, and Luis Rey. 2023. "The T6SS-Dependent Effector Re78 of Rhizobium etli Mim1 Benefits Bacterial Competition" Biology 12, no. 5: 678. https://doi.org/10.3390/biology12050678