Temporary Survival Increasing the Diversity of Culturable Heterotrophic Bacteria in the Newly Exposed Moraine at a Glacier Snout
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
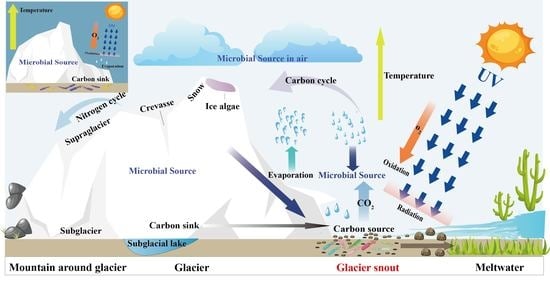
2.1. Site Description and Sample Collection
2.2. Analysis of the Physicochemical Properties of the Moraine
2.3. Isolation of Culturable Heterotrophic Bacteria using Distinct Culture Strategies
2.4. Identification of Bacterial Isolates through 16S rRNA Gene Sequencing
2.5. Growth Temperature Range and Radiation Resistance
2.6. Data Analysis
3. Results
3.1. Physicochemical Properties of the Moraine
3.2. Meteorological Characteristics of the Study Region
3.3. Abundance of Heterotrophic Bacteria and Phylogenetic Diversity of Strains at HT and LT
3.4. Comparison of Bacteria Cultured using Distinct Strategies
3.5. High Proportion of Isolated Potential Novel Taxa
3.6. Correlation between the Abundance of Heterotrophic Bacteria and Environmental Factors
3.7. Growth Temperature Range of Each Isolated Heterotrophic Bacterium
3.8. Radiation Resistance of Isolated Heterotrophic Bacteria
4. Discussion
4.1. Comparison of Bacteria Isolated from the Moraine of Laohugou Glacier No. 12 and Other Glacial Niches
4.2. Possible Effects of Temperature Changes on Bacteria Inhabiting the Newly Exposed Moraine
4.3. Temporary Survival and Potential Roles of Bacteria in Response to Different Environmental Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oerlemans, J. Quantifying global warming from the retreat of glaciers. Science 1994, 264, 243–245. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Huang, X.; Zhou, X.; Xiang, Q.; Zhao, K.; Yu, X.; Chen, Q.; Jiang, H.; Nyima, T.; Gao, X.; et al. Variation in denitrifying bacterial communities along a primary succession in the Hailuogou Glacier retreat area, China. PeerJ 2019, 7, e7356. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Gao, H.; Luo, W.; Elser, J.J. C:N:P stoichiometry in six distinct habitats of a glacier terminus in the source area of the Yangtze River. Biogeochemistry 2022, 158, 181–194. [Google Scholar] [CrossRef]
- Sigler, W.V.; Zeyer, J. Colony-forming analysis of bacterial community succession in deglaciated soils indicates pioneer stress-tolerant opportunists. Microb. Ecol. 2004, 48, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Zumsteg, A.; Luster, J.; Göransson, H.; Smittenberg, R.H.; Brunner, I.; Bernasconi, S.M.; Zeyer, J.; Frey, B. Bacterial, archaeal and fungal succession in the forefield of a receding glacier. Microb. Ecol. 2012, 63, 552–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Zhang, Y.; Chen, H.; Wang, Y.; Cao, F.; Sun, W.; Qi, X.; Zhao, Y.; Xu, F. Soil Properties and Microbial Diversity at the Frontier of Laohugou Glacier Retreat in Qilian Mountains. Curr. Microbiol. 2020, 77, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Wynn-Williams, D.D. Microbial processes and initial stabilization of fellfield soil. In Primary Succession on Land; Miles, J., Walton, D.W.H., Eds.; Blackwell Scientific: Hoboken, NJ, USA, 1993. [Google Scholar]
- Kong, W.; Liu, J.; Ji, M.; Yue, L.; Kang, S.; Morgan-Kiss, R.M. Autotrophic microbial community succession from glacier terminus to downstream waters on the Tibetan Plateau. FEMS Microbiol. Ecol. 2019, 95, fiz074. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, W.; Zheng, G.; Zhang, G.; Ma, X.; Xu, W.; Ali, B.; Rafiq, M. Diversity of Prokaryotic Communities Indigenous to Acid Mine Drainage and Related Rocks from Baiyin Open-Pit Copper Mine Stope, China. Geomicrobiol. J. 2018, 35, 580–600. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Tiwari, P.; Talukdar, G.; Rawat, G.S. Shifts in Bacterial Community Composition and Functional Traits at Different Time Periods Post-deglaciation of Gangotri Glacier, Himalaya. Curr. Microbiol. 2022, 79, 91. [Google Scholar] [CrossRef]
- Sajjad, W.; Ali, B.; Bahadur, A.; Ghimire, P.S.; Kang, S. Bacterial Diversity and Communities Structural Dynamics in Soil and Meltwater Runoff at the Frontier of Baishui Glacier No.1, China. Microb. Ecol. 2021, 81, 370–384. [Google Scholar] [CrossRef]
- Stibal, M.; Hasan, F.; Wadham, J.L.; Sharp, M.J.; Anesio, A.M. Prokaryotic diversity in sediments beneath two polar glaciers with contrasting organic carbon substrates. Extrem. Life Extrem. Cond. 2012, 16, 255–265. [Google Scholar] [CrossRef]
- Margesin, R.; Collins, T. Microbial ecology of the cryosphere (glacial and permafrost habitats): Current knowledge. Appl. Microbiol. Biotechnol. 2019, 103, 2537–2549. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Srinivas, T.N.; Pindi, P.K.; Kishore, K.H.; Begum, Z.; Singh, P.K.; Singh, A.K.; Pratibha, M.S.; Yasala, A.K.; Reddy, G.S.; et al. Bacterial biodiversity from Roopkund Glacier, Himalayan Mountain ranges, India. Extrem. Life Extrem. Cond. 2010, 14, 377–395. [Google Scholar] [CrossRef]
- Wilhelm, L.; Singer, G.A.; Fasching, C.; Battin, T.J.; Besemer, K. Microbial biodiversity in glacier-fed streams. ISME J. 2013, 7, 1651–1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.L.; Hou, S.G.; le Baoge, R.; Li, Z.G.; Xu, H.; Liu, Y.P.; Du, W.T.; Liu, Y.Q. Differences in Bacterial Diversity and Communities Between Glacial Snow and Glacial Soil on the Chongce Ice Cap, West Kunlun Mountains. Sci. Rep. 2016, 6, 36548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, B.; Sajjad, W.; Ghimire, P.S.; Shengyun, C.; Minghui, W.; Kang, S. Culture-dependent diversity of bacteria from Laohugou glacier, Qilian Mts., China and their resistance against metals. J. Basic Microbiol. 2019, 59, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hou, S.; Qin, X.; Du, W.; Liang, F.; Li, Z. Preliminary Study on Effects of Glacial Retreat on the Dominant Glacial Snow Bacteria in Laohugou Glacier No. 12. Geomicrobiol. J. 2014, 32, 113–118. [Google Scholar] [CrossRef]
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J.Z. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Microbiol. 2015, 13, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Pianka, E.R.J.T.A.N. On r- and K-Selection. Am. Nat. 1970, 104, 592–597. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, T.; Jiao, N.; Kang, S.; Huang, S.; Li, Q.; Wang, K.; Liu, X. Culturable bacteria in glacial meltwater at 6350 m on the East Rongbuk Glacier, Mount Everest. Extrem. Life Extrem. Cond. 2009, 13, 89–99. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Drake, H.L.; Gößner, A.S.; Daniel, S.L. Old Acetogens, New Light. Ann. N. Y. Acad. Sci. 2008, 1125, 100–128. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Dong, Z.; Kang, S.; Zong, C.; Rostami, M.; Shao, Y. Atmospheric deposition and contamination of trace elements in snowpacks of mountain glaciers in the northeastern Tibetan Plateau. Sci. Total Environ. 2019, 689, 754–764. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, T.; Guo, J.; Wu, M.; Yang, R.; Chen, X.; Wu, X.; Zhang, W.; Kang, S.; Liu, G.; et al. Microbial mercury methylation profile in terminus of a high-elevation glacier on the northern boundary of the Tibetan Plateau. Sci. Total Environ. 2020, 708, 135226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, T.; Li, J.; Wu, M.; Liu, G.; Zhang, W.; Zhang, B.; Zhang, S.; Zhang, G. High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert. Biology 2022, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Qin, D.; Chen, J.; Qin, X.; Ren, J.; Cui, X.; Du, Z.; Kang, S. Physicochemical impacts of dust particles on alpine glacier meltwater at the Laohugou Glacier basin in western Qilian Mountains, China. Sci. Total Environ. 2014, 493, 930–942. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics; John Wiley and Sons: New York, NY, USA, 1991. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; de Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.; Cui, X.; Xu, Y.; Hu, S.; Zhao, Y.; Zhang, W.; Liu, G.; Zhang, G. Sphingomonas radiodurans sp. nov., a novel radiation-resistant bacterium isolated from the north slope of Mount Everest. Int. J. Syst. Evol. Microbiol. 2022, 72, 005312. [Google Scholar] [CrossRef]
- Rew, D.A. Deinococcus radiodurans. Eur. J. Surg. Oncol. 2003, 29, 557–558. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef] [PubMed]
- ter Braak, C.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software of Ordination; Version 5.0; Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Li, Q.; Liu, Y.; Gu, Y.; Guo, L.; Huang, Y.; Zhang, J.; Xu, Z.; Tan, B.; Zhang, L.; Chen, L.; et al. Ecoenzymatic stoichiometry and microbial nutrient limitations in rhizosphere soil along the Hailuogou Glacier forefield chronosequence. Sci. Total Environ. 2020, 704, 135413. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; de Carvalho, C.C.C.R.; Stevenson, A.; Grant, I.R.; Hallsworth, J.E. Extraordinary solute-stress tolerance contributes to the environmental tenacity of mycobacteria. Environ. Microbiol. Rep. 2015, 7, 746–764. [Google Scholar] [CrossRef]
- Johnson, S.S.; Hebsgaard, M.B.; Christensen, T.R.; Mastepanov, M.; Nielsen, R.; Munch, K.; Brand, T.; Gilbert, M.T.; Zuber, M.T.; Bunce, M.; et al. ancient bacteria show evidence of DNA repair. Proc. Natl. Acad. Sci. USA 2007, 104, 14401–14405. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.; Sangwan, N.; Lal, R.; Vakhlu, J. Bacterial diversity of Drass, cold desert in Western Himalaya, and its comparison with Antarctic and Arctic. Arch. Microbiol. 2015, 197, 851–860. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [Green Version]
- Pearce, D.A.; Bridge, P.D.; Hughes, K.A.; Sattler, B.; Psenner, R.; Russell, N.J. Microorganisms in the atmosphere over Antarctica. FEMS Microbiol. Ecol. 2009, 69, 143–157. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Bhat, S.; Fraser, M.P. Characterization of saccharides and other organic compounds in fine particles and the use of saccharides to track primary biologically derived carbon sources. Atmos. Environ. 2010, 44, 724–732. [Google Scholar] [CrossRef]
- Zhang, S.; Hou, S.; Baoge, R.; Xu, H.; Liu, Y.; Li, Z. Difference of community structure among culturable bacteria in different glacial samples on Chongce Ice Cap. Wei Sheng Wu Xue Bao Acta Microbiol. Sin. 2016, 56, 708–718. [Google Scholar]
- Nemergut, D.R.; Anderson, S.P.; Cleveland, C.C.; Martin, A.P.; Miller, A.E.; Seimon, A.; Schmidt, S.K. Microbial community succession in an unvegetated, recently deglaciated soil. Microb. Ecol. 2007, 53, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, M.; Anderson, S.P.; Sharp, M.; Foght, J.; Lanoil, B.D. Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl. Environ. Microbiol. 2005, 71, 6986–6997. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Lawrence, J.R.; Sanschagrin, S.; Waiser, M.J.; Korber, D.R.; Greer, C.W. Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl. Environ. Microbiol. 2012, 78, 7626–7637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yao, T.; Jiao, N.; Tian, L.; Hu, A.; Yu, W.; Li, S. Microbial diversity in the snow, a moraine lake and a stream in Himalayan glacier. Extrem. Life Extrem. Cond. 2011, 15, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Bahadur, A.; Zhang, W.; Sajjad, W.; Nasir, F.; Zhang, G.; Liu, G.; Chen, T. Bacterial diversity patterns of desert dunes in the northeastern Qinghai-Tibet Plateau, China. Arch. Microbiol. 2021, 203, 2809–2823. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, H.H.; Yuan, M.; Yao, Q.; Tang, R.; Lin, M.; Yang, S.Z.; Li, Z.K.; Chen, M. Microbacterium radiodurans sp. nov., a UV radiation-resistant bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 2665–2670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippidou, S.; Junier, T.; Wunderlin, T.; Lo, C.C.; Li, P.E.; Chain, P.S.; Junier, P. Under-detection of endospore-forming Firmicutes in metagenomic data. Comput. Struct. Biotechnol. J. 2015, 13, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Liu, J.; Kong, W.; Xia, P.; Zhu, C.; Li, X. Prokaryotic Community Succession in Bulk and Rhizosphere Soils Along a High-Elevation Glacier Retreat Chronosequence on the Tibetan Plateau. Front. Microbiol. 2021, 12, 736407. [Google Scholar] [CrossRef]
- Bakermans, C. Determining the Limits of Microbial Life at Subzero Temperatures. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 21–38. [Google Scholar]
- Raymond-Bouchard, I.; Tremblay, J.; Altshuler, I.; Greer, C.W.; Whyte, L.G. Comparative Transcriptomics of Cold Growth and Adaptive Features of a Eury- and Steno-Psychrophile. Front. Microbiol. 2018, 9, 1565. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.D. Nocardioides furvisabuli sp. nov., isolated from black sand. Int. J. Syst. Evol. Microbiol. 2007, 57, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Carro, L.; Castro, J.F.; Razmilic, V.; Nouioui, I.; Pan, C.; Igual, J.M.; Jaspars, M.; Goodfellow, M.; Bull, A.T.; Asenjo, J.A.; et al. Uncovering the potential of novel micromonosporae isolated from an extreme hyper-arid Atacama Desert soil. Sci. Rep. 2019, 9, 4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reice, S.R. Experimental disturbance and the maintenance of species diversity in a stream community. Oecologia 1985, 67, 90–97. [Google Scholar] [CrossRef]
- Petraitis, P.S.; Latham, R.E.; Niesenbaum, R.A. The Maintenance of Species Diversity by Disturbance. Q. Rev. Biol. 1989, 64, 393–418. [Google Scholar] [CrossRef]
- O’Malley, M.A. ‘Everything is everywhere: But the environment selects’: Ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Biol. Biomed. Sci. 2008, 39, 314–325. [Google Scholar] [CrossRef]
- Ortiz-Álvarez, R.; Fierer, N.; de Los Ríos, A.; Casamayor, E.O.; Barberán, A. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J. 2018, 12, 1658–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Menor, E.; Gimeno-Valero, H.; Pascual, J.; Peretó, J.; Porcar, M. High Culturable Bacterial Diversity from a European Desert: The Tabernas Desert. Front. Microbiol. 2020, 11, 583120. [Google Scholar] [CrossRef] [PubMed]
- Dube, J.P.; Valverde, A.; Steyn, J.M.; Cowan, D.A.; van der Waals, J.E. Differences in Bacterial Diversity, Composition and Function due to Long-Term Agriculture in Soils in the Eastern Free State of South Africa. Diversity 2019, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, Y.; Li, J.; He, Q.; Zhu, H.; Bing, H. Energetic supply regulates heterotrophic nitrogen fixation along a glacial chronosequence. Soil Biol. Biochem. 2021, 154, 108150. [Google Scholar] [CrossRef]
- Dieser, M.; Broemsen, E.L.; Cameron, K.A.; King, G.M.; Achberger, A.; Choquette, K.; Hagedorn, B.; Sletten, R.; Junge, K.; Christner, B.C. Molecular and biogeochemical evidence for methane cycling beneath the western margin of the Greenland Ice Sheet. ISME J. 2014, 8, 2305–2316. [Google Scholar] [CrossRef]
- Nash, M.V.; Anesio, A.M.; Barker, G.; Tranter, M.; Varliero, G.; Eloe-Fadrosh, E.A.; Nielsen, T.; Turpin-Jelfs, T.; Benning, L.G.; Sánchez-Baracaldo, P. Metagenomic insights into diazotrophic communities across Arctic glacier forefields. FEMS Microbiol. Ecol. 2018, 94, fiy114. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Hall, J.; Ryan, K. Low salinity and high-level UV-B radiation reduce single-cell activity in antarctic sea ice bacteria. Appl. Environ. Microbiol. 2009, 75, 7570–7573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, P.B. Microbial genesis, life and death in glacial ice. Can. J. Microbiol. 2009, 55, 1–11. [Google Scholar] [CrossRef]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef]
- Coppola, D.; Verde, C.; Giordano, D. Isolation of UV-Resistant Marine Bacteria by UV-C Assays. In Marine Genomics, Methods Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2498, pp. 293–305. [Google Scholar] [CrossRef]
- Hellingwerf, K.J.; Crielaard, W.; Hoff, W.D.; Matthijs, H.C.; Mur, L.R.; van Rotterdam, B.J. Photobiology of bacteria. Antonie Leeuwenhoek 1994, 65, 331–347. [Google Scholar] [CrossRef] [Green Version]
- Li, I.V.; Terekhova, L.P.; Gapochka, M.G. Isolation of actinomycetes from soil using extremely high frequency radiation. Mikrobiologiia 2002, 71, 119–122. [Google Scholar] [PubMed]
- Liu, Y.; Yao, T.; Jiao, N.; Kang, S.; Zeng, Y.; Huang, S. Microbial community structure in moraine lakes and glacial meltwaters, Mount Everest. FEMS Microbiol. Lett. 2006, 265, 98–105. [Google Scholar] [CrossRef] [Green Version]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Zagorodnov, V.; Sandman, K.; Reeve, J.N. Recovery and Identification of Viable Bacteria Immured in Glacial Ice. Icarus 2000, 144, 479–485. [Google Scholar] [CrossRef]
- Foght, J.; Aislabie, J.; Turner, S.; Brown, C.E.; Ryburn, J.; Saul, D.J.; Lawson, W. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb. Ecol. 2004, 47, 329–340. [Google Scholar] [CrossRef]
- Vitousek, P.M.; White, P.S. Process Studies in Succession. In Forest Succession: Concepts and Application; West, D.C., Shugart, H.H., Botkin, D.B., Eds.; Springer New York: New York, NY, USA, 1981; pp. 267–276. [Google Scholar]
- Andersson, D.I.; Levin, B.R. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 1999, 2, 489–493. [Google Scholar] [CrossRef]








 | HT | Blastococcus Geodermatophilus Marmoricola Microbacterium Micrococcus Micromonospora Mycolicibacterium | Promicromonospora Pseudonocardia Rhodococcus Streptomyces Phenylobacterium Paeniglutamicibacter Sphingorhabdus | Caulobacter Dankookia Erythrobacter Microvirga Pararhizobium Roseomonas Longivirga | Bacillus Neobacillus Peribacillus Belnapia Rudaea Bosea Spirosoma |
| Shared | Mycetocola Pseudarthrobacter Hymenobacter Modestobacter | Methylobacterium Noviherbaspirillum Brevundimonas Cellulomonas | Deinococcus Acidovorax Sphingomonas Arthrobacter | Cryobacterium Clavibacter Agria Nocardioides | |
| LT | Larkinella Aureimonas Marisediminicola | Phyllobacterium Polymorphobacter Flavobacterium | Skermanella Polaromonas Knoellia | Massilia Plantibacter Adhaeribacter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xu, Y.; Cui, X.; Zhang, B.; Wang, X.; Qin, X.; Wang, J.; Li, Y.; Zhang, W.; Liu, G.; et al. Temporary Survival Increasing the Diversity of Culturable Heterotrophic Bacteria in the Newly Exposed Moraine at a Glacier Snout. Biology 2022, 11, 1555. https://doi.org/10.3390/biology11111555
Liu Y, Xu Y, Cui X, Zhang B, Wang X, Qin X, Wang J, Li Y, Zhang W, Liu G, et al. Temporary Survival Increasing the Diversity of Culturable Heterotrophic Bacteria in the Newly Exposed Moraine at a Glacier Snout. Biology. 2022; 11(11):1555. https://doi.org/10.3390/biology11111555
Chicago/Turabian StyleLiu, Yang, Yeteng Xu, Xiaowen Cui, Binglin Zhang, Xinyue Wang, Xiang Qin, Jinxiu Wang, Yanzhao Li, Wei Zhang, Guangxiu Liu, and et al. 2022. "Temporary Survival Increasing the Diversity of Culturable Heterotrophic Bacteria in the Newly Exposed Moraine at a Glacier Snout" Biology 11, no. 11: 1555. https://doi.org/10.3390/biology11111555