Acinetobacter baumannii Survival under Infection-Associated Stresses Depends on the Expression of Resistance–Nodulation–Division and Major Facilitator Superfamily Efflux Pumps
Abstract
:1. Introduction
2. Results
2.1. MFS Pumps Do Not Contribute to Antibiotic Efflux
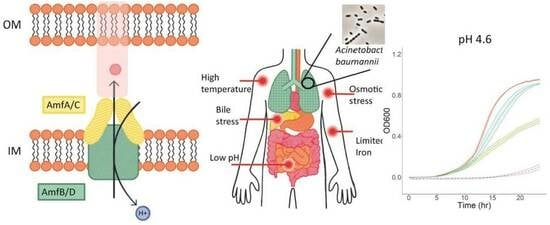
2.2. Both AmfAB and AmfCD Pumps Are Important for Growth under Acidic Conditions, but AmfCD also Contributes to Survival under Other Stresses
2.3. RND Pumps Dominate the Growth Phenotypes of A. baumannii under Virulence-Related Stress Conditions
2.4. Principal Component Analysis Reveals Differences between the Strains
2.5. Efflux Pump Deletions and Overproducers Modify the Permeability Barrier of A. baumannii
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Deletion of A1S_1772–1773, A1S_1799–1800 Operons
4.3. Construction of Plasmids for Overproduction of Efflux Pumps
4.4. Drug Susceptibility Assay (Minimal Inhibitory Concentration (MIC) Determination)
4.5. NPN Uptake
4.6. Bacterial Growth with Stress Exposures
4.7. Protein Expression and Analyses
4.8. Clustering
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Houang, E.T.; Sormunen, R.T.; Lai, L.; Chan, C.Y.; Leong, A.S. Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J. Clin. Pathol. 1998, 51, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert. Rev. Anti Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.M.P.D.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022.
- Kornelsen, V.; Kumar, A. Update on Multidrug Resistance Efflux Pumps in Acinetobacter spp. Antimicrob. Agents Chemother. 2021, 65, e0051421. [Google Scholar] [CrossRef] [PubMed]
- Rybenkov, V.V.; Zgurskaya, H.I.; Ganguly, C.; Leus, I.V.; Zhang, Z.; Moniruzzaman, M. The Whole Is Bigger than the Sum of Its Parts: Drug Transport in the Context of Two Membranes with Active Efflux. Chem. Rev. 2021, 121, 5597–5631. [Google Scholar] [CrossRef]
- Geisinger, E.; Huo, W.; Hernandez-Bird, J.; Isberg, R.R. Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu. Rev. Microbiol. 2019, 73, 481–506. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Courvalin, P.; Grillot-Courvalin, C. RND-Type Efflux Pumps in Multidrug-Resistant Clinical Isolates of Acinetobacter baumannii: Major Role for AdeABC Overexpression and AdeRS Mutations. Antimicrob. Agents Chemother. 2013, 57, 2989–2995. [Google Scholar] [CrossRef]
- Du, D.; van Veen, H.W.; Murakami, S.; Pos, K.M.; Luisi, B.F. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr. Opin. Struct. Biol. 2015, 33, 76–91. [Google Scholar] [CrossRef]
- Leus, I.V.; Adamiak, J.; Trinh, A.N.; Smith, R.D.; Smith, L.; Richardson, S.; Ernst, R.K.; Zgurskaya, H.I. Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol. Microbiol. 2020, 114, 1049–1065. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Balloy, V.; Fiette, L.; Chignard, M.; Courvalin, P.; Grillot-Courvalin, C. Contribution of the Ade Resistance-Nodulation-Cell Division-Type Efflux Pumps to Fitness and Pathogenesis of Acinetobacter baumannii. mBio 2016, 7, e00697-16. [Google Scholar] [CrossRef] [PubMed]
- Leus, I.V.; Weeks, J.W.; Bonifay, V.; Smith, L.; Richardson, S.; Zgurskaya, H.I. Substrate Specificities and Efflux Efficiencies of RND Efflux Pumps of Acinetobacter baumannii. J. Bacteriol. 2018, 200, e00049-18. [Google Scholar] [CrossRef]
- Jiang, J.H.; Hassan, K.A.; Begg, S.L.; Rupasinghe, T.W.T.; Naidu, V.; Pederick, V.G.; Khorvash, M.; Whittall, J.J.; Paton, J.C.; Paulsen, I.T.; et al. Identification of Novel Acinetobacter baumannii Host Fatty Acid Stress Adaptation Strategies. mBio 2019, 10, e02056-02018. [Google Scholar] [CrossRef] [PubMed]
- Palma, V.; Gutiérrez, M.S.; Vargas, O.; Parthasarathy, R.; Navarrete, P. Methods to Evaluate Bacterial Motility and Its Role in Bacterial–Host Interactions. Microorganisms 2022, 10, 563. [Google Scholar] [PubMed]
- Lomovskaya, O.; Lewis, K. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 1992, 89, 8938–8942. [Google Scholar] [CrossRef] [PubMed]
- Avican, K.; Aldahdooh, J.; Togninalli, M.; Mahmud, A.K.M.F.; Tang, J.; Borgwardt, K.M.; Rhen, M.; Fällman, M. RNA atlas of human bacterial pathogens uncovers stress dynamics linked to infection. Nat. Commun. 2021, 12, 3282. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Adamiak, J.W.; Leus, I.V. Making sense of drug-efflux transporters in the physiological environment. Curr. Opin. Microbiol. 2022, 69, 102179. [Google Scholar] [CrossRef]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar] [CrossRef]
- Zeng, S.; Shi, Q.; Liu, Y.; Li, M.; Lin, D.; Zhang, S.; Li, Q.; Pu, J.; Shen, C.; Huang, B.; et al. The small RNA PrrH of Pseudomonas aeruginosa regulates hemolysis and oxidative resistance in bloodstream infection. Microb. Pathog. 2023, 180, 106124. [Google Scholar] [CrossRef]
- Liao, C.-H.; Chen, W.-C.; Li, L.-H.; Lin, Y.-T.; Pan, S.-Y.; Yang, T.-C. AmpR of Stenotrophomonas maltophilia is involved in stenobactin synthesis and enhanced β-lactam resistance in an iron-depleted condition. J. Antimicrob. Chemother. 2020, 75, 3544–3551. [Google Scholar] [CrossRef]
- Wu, C.J.; Chen, Y.; Li, L.H.; Wu, C.M.; Lin, Y.T.; Ma, C.H.; Yang, T.C. Roles of SmeYZ, SbiAB, and SmeDEF Efflux Systems in Iron Homeostasis of Stenotrophomonas maltophilia. Microbiol. Spectr. 2022, 10, e0244821. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, G.; Leus, I.V.; Weeks, J.W.; Wolloscheck, D.; Rybenkov, V.V.; Zgurskaya, H.I. Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. mBio 2017, 8, e01172-17. [Google Scholar] [CrossRef] [PubMed]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R.; et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, K.; Brameyer, S.; Jung, K. Bacterial acid stress response: From cellular changes to antibiotic tolerance and phenotypic heterogeneity. Curr. Opin. Microbiol. 2023, 75, 102367. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Fanelli, G.; Pasqua, M.; Colonna, B.; Prosseda, G.; Grossi, M. Expression Profile of Multidrug Resistance Efflux Pumps during Intracellular Life of Adherent-Invasive Escherichia coli Strain LF82. Front. Microbiol. 2020, 11, 1935. [Google Scholar] [CrossRef]
- Pasqua, M.; Grossi, M.; Scinicariello, S.; Aussel, L.; Barras, F.; Colonna, B.; Prosseda, G. The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep. 2019, 9, 2906. [Google Scholar] [CrossRef] [PubMed]
- Distel, J.S.; Di Venanzio, G.; Mackel, J.J.; Rosen, D.A.; Feldman, M.F. Replicative Acinetobacter baumannii strains interfere with phagosomal maturation by modulating the vacuolar pH. PLoS Pathog. 2023, 19, e1011173. [Google Scholar] [CrossRef] [PubMed]
- Lewinson, O.; Padan, E.; Bibi, E. Alkalitolerance: A biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA 2004, 101, 14073–14078. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.A.; Skurnik, M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 2000, 37, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-F.; Lin, Y.-Y.; Lan, C.-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 2017, 55, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Tseng, W.; Guina, T.; Miller, S.I.; Nikaido, H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J. Bacteriol. 2007, 189, 7213–7222. [Google Scholar] [CrossRef] [PubMed]
- Van Riet, S.; Tadesse, W.; Mortier, J.; Schlegel, S.; Simoens, K.; Bernaerts, K.; Dal Co, A.; Aertsen, A. Heterogeneity and Evolutionary Tunability of Escherichia coli Resistance against Extreme Acid Stress. Microbiol. Spectr. 2022, 10, e0375722. [Google Scholar] [CrossRef]
- Bearson, S.; Bearson, B.; Foster, J.W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997, 147, 173–180. [Google Scholar] [CrossRef]
- Ren, X.; Palmer, L.D. Acinetobacter Metabolism in Infection and Antimicrobial Resistance. Infect. Immun. 2023, 91, e00433-22. [Google Scholar] [CrossRef]
- Whittle, E.E.; McNeil, H.E.; Trampari, E.; Webber, M.; Overton, T.W.; Blair, J.M.A. Efflux Impacts Intracellular Accumulation Only in Actively Growing Bacterial Cells. mBio 2021, 12, e0260821. [Google Scholar] [CrossRef]
- Tucker, A.T.; Nowicki, E.M.; Boll, J.M.; Knauf, G.A.; Burdis, N.C.; Trent, M.S.; Davies, B.W. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 2014, 5, e01313–e01314. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, G.; Wolloscheck, D.; Weeks, J.W.; Croft, C.; Rybenkov, V.V.; Zgurskaya, H.I. Breaking the Permeability Barrier of Escherichia coli by Controlled Hyperporination of the Outer Membrane. Antimicrob. Agents Chemother. 2016, 60, 7372–7381. [Google Scholar] [CrossRef] [PubMed]
- Ram, Y.; Dellus-Gur, E.; Bibi, M.; Karkare, K.; Obolski, U.; Feldman, M.W.; Cooper, T.F.; Berman, J.; Hadany, L. Predicting microbial growth in a mixed culture from growth curve data. Proc. Natl. Acad. Sci. USA 2019, 116, 14698–14707. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
- Leus, I.V.; Adamiak, J.; Chandar, B.; Bonifay, V.; Zhao, S.; Walker, S.S.; Squadroni, B.; Balibar, C.J.; Kinarivala, N.; Standke, L.C.; et al. Functional Diversity of Gram-Negative Permeability Barriers Reflected in Antibacterial Activities and Intracellular Accumulation of Antibiotics. Antimicrob. Agents Chemother. 2023, 67, e0137722. [Google Scholar] [CrossRef] [PubMed]
- Fabre, L.; Ntreh, A.T.; Yazidi, A.; Leus, I.V.; Weeks, J.W.; Bhattacharyya, S.; Ruickoldt, J.; Rouiller, I.; Zgurskaya, H.I.; Sygusch, J. A “Drug Sweeping” State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa. Structure 2021, 29, 261–274.e266. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Damron, F.H.; McKenney, E.S.; Barbier, M.; Liechti, G.W.; Schweizer, H.P.; Goldberg, J.B. Construction of mobilizable mini-Tn7 vectors for bioluminescent detection of gram-negative bacteria and single-copy promoter lux reporter analysis. Appl. Environ. Microbiol. 2013, 79, 4149–4153. [Google Scholar] [CrossRef]
- Amin, I.M.; Richmond, G.E.; Sen, P.; Koh, T.H.; Piddock, L.J.V.; Chua, K.L. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol. 2013, 13, 158. [Google Scholar] [CrossRef]




| Strain | Ery | Novo | Cipro | Tet | Azi | Cm | SDS | Gm | Zeo |
|---|---|---|---|---|---|---|---|---|---|
| AbWT | 10 | 10–20 | 0.125–0.25 | 0.25–0.5 | 0.63–1.25 | 64 | >1024 | 4–8 | 8 |
| ∆AmfAB | 5 | 10 | 0.125–0.25 | 0.5 | 0.63–1.25 | 64 | >1024 | 8 | 8 |
| ∆AmfCD | 5–10 | 5–10 | 0.125 | 0.25–0.5 | 1.25 | 64 | >1024 | 8 | 8 |
| ∆2 | 10 | 10 | 0.125 | 0.25 | 1.25 | 32 | ≥1024 | R | 8 |
| AbWT(pAmfAB) | 2.5–5 | 5–10 | 0.125 | 0.5–1 | 0.31–0.63 | 64 | >1024 | 4 | 2 |
| AbWT(pAmfCD) | 2.5–5 | 10–20 | 0.125 | 0.5 | 0.63 | 64 | >1024 | 4 | 4 |
| ∆2(pAmfAB) | 10 | 5–10 | 0.0625–0.125 | 0.5 | 0.63–1.25 | 32 | >1024 | R | 4–8 |
| ∆2(pAmfCD) | 5 | 5–10 | 0.0625–0.125 | 0.25 | 1.25 | 32 | >1024 | R | 16 |
| Ab∆3 | 1.25 | 0.08 | 0.016–0.031 | 0.031–0.063 | 0.31–0.63 | 16 | 16 | 8 | 1 |
| ∆3∆AmfAB | 1.25 | 0.08 | 0.008–0.016 | 0.031 | 0.63 | 8–16 | 16 | 8 | 1 |
| ∆3∆AmfCD | 1.25 | 0.08 | 0.016 | 0.031 | 0.63 | 8–16 | 16 | 8 | 1–2 |
| ∆5 | 2.5 | 0.08 | 0.016 | 0.031 | 0.31–0.63 | 8 | 16 | R | 1–2 |
| ∆3(pAmfAB) | 5 | 0.04 | 0.016 | 0.016–0.031 | 0.31 | 8 | 16 | 8 | 1–2 |
| ∆3(pAmfCD) | 2.5–5 | 0.04 | 0.016 | 0.016 | 0.31–0.63 | 8 | 16 | 8 | 0.5–1 |
| ∆5(pAmfAB) | 5 | 0.04 | 0.016 | 0.016–0.031 | 0.63 | 8 | 16 | R | 0.25–0.5 |
| ∆5(pAmfCD) | 2.5–5 | 0.04 | 0.016 | 0.016–0.031 | 0.63 | 8 | 16 | R | 0.5–1 |
| Type of Stress | Stress Exposure Agent in LB Broth or LB Broth + 1% L-Arabinose |
|---|---|
| No stress control | LB broth, pH 7.2; 37 °C |
| Acidic stress | LB broth adjusted to pH 4.6 with HCl |
| Osmotic stress | LB broth supplemented with 0.5 M NaCl |
| Bile stress | LB broth supplemented with 0.5% bile salts (Millipore (Darmstadt, Germany), ~50% sodium cholate and ~50% sodium deoxycholate) |
| Low iron | LB broth supplemented with 125 μM or 250 μM 2,2-dipyridyl |
| Temperature | LB broth, pH 7.2; 41 °C |
| Strains | No Stress | Acid | Osmotic | Bile | Iron Depletion | High Temperature |
|---|---|---|---|---|---|---|
| 37 °C, pH 7.2 | pH 4.6 | 0.5 M NaCl | 0.5% Bile Salts | 2,2′-Dipyridyl (250 mM) | 41 °C | |
| WT | 12.13 ± 0.96 | 11.71 ± 2.91 | 3.48 ± 1 | 4.16 ± 2.19 | 3.66 ± 2.46 | 12.1 ± 4.21 |
| ∆AmfAB | 12.34 ± 1.17 | 8.26 ± 2.1 | 3.89 ± 0.64 | 4.65 ± 1.47 | 4.13 ± 2.25 | 12.92 ± 2.79 |
| ∆AmfCD | 10.16 ± 0.86 | 4.59 ± 2.48 | 2.92 ± 0.26 | 2.6 ± 0.81 | 2.78 ± 0.54 | 7.96 ± 0.94 |
| ∆2 | 9.42 ± 0.92 | 3.59 ± 0.9 | 3.42 ± 0.36 | 2.45 ± 0.77 | 2.36 ± 1.11 | 9.98 ± 2.92 |
| ∆3 | 10.81 ± 1.43 | 1.67 ± 0.81 | 2.59 ± 0.65 | 0.46 ± 0.28 | 1.69 ± 1.38 | 10.04 ± 3.46 |
| ∆3∆AmfAB | 10.65 ± 0.67 | 1.44 ± 0.71 | 3.14 ± 0.48 | NG | 1.12 ± 0.45 | 8.91 ± 3.05 |
| ∆3∆AmfCD | 9.89 ± 0.87 | 2.57 ± 1.27 | 2.97 ± 0.44 | NG | 1.94 ± 0.59 | 10.17 ± 2.9 |
| ∆5 | 9.16 ± 0.9 | 2.06 ± 1.21 | 2.99 ± 0.51 | NG | 1.14 ± 0.76 | 8.98 ± 2.23 |
| Strains | No Stress | Acid | Osmotic | Bile | Iron Depletion | High Temperature |
|---|---|---|---|---|---|---|
| 37 °C, pH 7.2 | pH 4.6 | 0.5 M NaCl | 0.5% DOC | 2,2′-Dipyridyl (125 mM) | 41 °C | |
| WT | 11.89 ± 1.47 | 9.39 ± 0.61 | 5.94 ± 2.36 | 4.78 ± 0.58 | 9.93 ± 5.7 | 9.78 ± 3.03 |
| WT(pAmfAB) | 5.21 ± 1.23 | 4.85 ± 1.04 | 1.61 ± 0.12 | 2.62 ± 0.24 | 5.41 ± 0.08 | 5.33 ± 0.57 |
| WT(pAmfCD) | 11.83 ± 0.69 | 7.91 ± 1.9 | 3.47 ± 0.39 | 3.77 ± 0.27 | 8.67 ± 0.13 | 11.95 ± 0.57 |
| D2 | 8.19 ± 0.53 | 0.16 ± 0.21 | 5.04 ± 0.49 | 2.47 ± 0.17 | 4.28 ± 0.17 | 8.48 ± 0.59 |
| D2(pAmfAB) | 6.12 ± 0.7 | 4.3 ± 0.7 | 4.33 ± 0.44 | 1.3 ± 0.32 | 4 ± 0.67 | 8.15 ± 0.12 |
| D2(pAmfCD) | 10.95 ± 0.7 | 5.49 ± 0.96 | 5.43 ± 0.11 | 2.71 ± 1.1 | 7.96 ± 0.4 | 11.88 ± 0.21 |
| D3 | 10.62 ± 1.27 | 1.13 ± 0.13 | 3.19 ± 0.78 | NG | 7.52 ± 4.82 | 9.1 ± 2.14 |
| D3(pAmfAB) | 7.2 ± 1.78 | 0.08 ± 0.08 | 0.98 ± 0.62 | 0.39 ± 0.7 | 4.69 ± 0.01 | 7.58 ± 0.4 |
| D3(pAmfCD) | 9.65 ± 1.17 | 0.04 ± 0.07 | 2.32 ± 0.44 | 0.12 ± 0.31 | 4.96 ± 0.19 | 9.95 ± 0.91 |
| D5 | 8.01 ± 1.01 | NG | 4.81 ± 0.59 | 0.57 ± 0.42 | 4.45 ± 0.32 | 7.61 ± 0.4 |
| D5(pAmfAB) | 7.3 ± 1.08 | NG | 3.96 ± 0.59 | 0.62 ± 0.6 | 3.49 ± 0.69 | 8.73 ± 0.37 |
| D5(pAmfCD) | 8.07 ± 0.75 | NG | 4.65 ± 0.31 | 0.14 ± 0.35 | 4.04 ± 0.74 | 7.73 ± 0.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leus, I.V.; Olvera, M.; Adamiak, J.W.; Nguyen, L.L.; Zgurskaya, H.I. Acinetobacter baumannii Survival under Infection-Associated Stresses Depends on the Expression of Resistance–Nodulation–Division and Major Facilitator Superfamily Efflux Pumps. Antibiotics 2024, 13, 7. https://doi.org/10.3390/antibiotics13010007
Leus IV, Olvera M, Adamiak JW, Nguyen LL, Zgurskaya HI. Acinetobacter baumannii Survival under Infection-Associated Stresses Depends on the Expression of Resistance–Nodulation–Division and Major Facilitator Superfamily Efflux Pumps. Antibiotics. 2024; 13(1):7. https://doi.org/10.3390/antibiotics13010007
Chicago/Turabian StyleLeus, Inga V., Marcela Olvera, Justyna W. Adamiak, Lauren L. Nguyen, and Helen I. Zgurskaya. 2024. "Acinetobacter baumannii Survival under Infection-Associated Stresses Depends on the Expression of Resistance–Nodulation–Division and Major Facilitator Superfamily Efflux Pumps" Antibiotics 13, no. 1: 7. https://doi.org/10.3390/antibiotics13010007