Synergistic Antibacterial Effects of Amoxicillin and Gold Nanoparticles: A Therapeutic Option to Combat Antibiotic Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Bacterial Lines and Culture Conditions
2.1.2. Synthesis of 16-mph-16 Surfactant
2.1.3. Synthesis of Au@16-mph-16 Nanoparticles
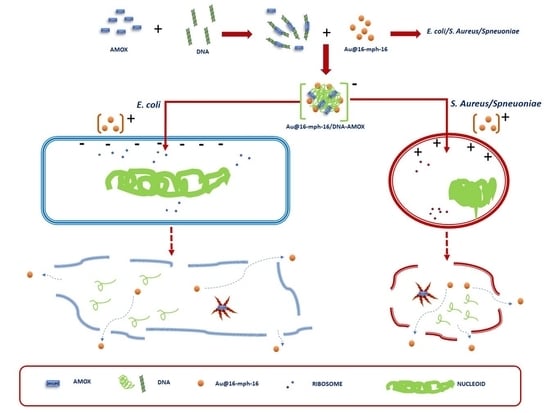
2.1.4. Synthesis of Au@16-mph-16/DNA-AMOX Nanosystem
2.2. Methods
2.2.1. DNA Fragmentation
2.2.2. UV/Vis Spectroscopy
2.2.3. Circular Dichroism Spectroscopy (CD)
2.2.4. Zeta Potential Measurements and Dynamic Light Scattering (DLS)
2.2.5. Transmission Electron Microscopy (TEM)
2.2.6. Energy-Dispersive Spectroscopy (EDS) Measurements
2.2.7. Gold Nanosystems’ Susceptibility Tests against Reference Strains
2.2.8. Evaluation of Nanoparticles and Nanosystems on Agar Plates
2.2.9. Preparation of Pellets for TEM and SEM Microscopes
3. Results and Discussion
3.1. Au@16-mph-16/DNA-AMOX Complex Formation and Conformational Changes in DNA/AMOX Complexes Induced by Au@16-mph-16 Cationic Nanoparticles
3.2. Stability, Charge, and Size of Au@16-mph-16 and Au@ 16-mph-16/DNA-AMOX Nanosystems and Release Kinetics
3.3. Results of Gold Nanosystems’ Susceptibility Tests against Gram+ and Gram− Reference Strains
3.4. Internalization of Au@16-mph-16 and Au@16-mph-16/DNA-AMOX Nanosystems
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 6 July 2023).
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022; World Health Organization: Geneva, Switzerland, 2022; ISBN 9789240062702. [Google Scholar]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Adisasmito, W.B.; Almuhairi, S.; Behravesh, C.B.; Bilivogui, P.; Bukachi, S.A.; Casas, N.; Becerra, N.C.; Charron, D.F.; Chaudhary, A.; Ciacci Zanella, J.R.; et al. One Health: A New Definition for a Sustainable and Healthy Future. PLoS Pathog. 2022, 18, e1010537. [Google Scholar] [CrossRef]
- Chapman, J.S.; Georgopapdakou, N.H. Routes of Quinolone Permeation in Escherichia Coli. Antimicrob. Agents Chemother. 1988, 32, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delcour, A.H. Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, K. Efflux-Mediated Antimicrobial Resistance. J. Antimicrob. Chemother. 2005, 56, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Huttner, A.; Bielicki, J.; Clements, M.N.; Frimodt-Møller, N.; Muller, A.E.; Paccaud, J.-P.; Mouton, J.W. Oral Amoxicillin and Amoxicillin–Clavulanic Acid: Properties, Indications and Usage. Clin. Microbiol. Infect. 2020, 26, 871–879. [Google Scholar] [CrossRef]
- Visentin, M.; Lenggenhager, D.; Gai, Z.; Kullak-Ublick, G.A. Drug-Induced Bile Duct Injury. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1498–1506. [Google Scholar] [CrossRef]
- Dhir, A.; Kular, H.; Elzagallaai, A.A.; Carleton, B.; Rieder, M.J.; Mak, R.; Wong, T. DRESS Induced by Amoxicillin-Clavulanate in Two Pediatric Patients Confirmed by Lymphocyte Toxicity Assay. Allergy Asthma Clin. Immunol. 2021, 17, 37. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Jang, J.; Hur, H.-G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia Coli: Ecology and Public Health Implications—A Review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [Green Version]
- Prieto Prieto, J.; García del Potro, M.; Gómez-Lus Centelles, M.L. Guía Para El Uso de Antibióticos En Atención Primaria. In Eficacia In Vivo, Eficacia In Vitro; Ediciones Doyma: Madrid, Spain, 1997; pp. 83–96. [Google Scholar]
- Tomasz, A. Antibiotic Resistance in Streptococcus Pneumoniae. Clin. Infect. Dis. 1997, 24, S85–S88. [Google Scholar] [CrossRef] [PubMed]
- Henriques-Normark, B.; Tuomanen, E.I. The Pneumococcus: Epidemiology, Microbiology, and Pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, P.C. Resistance among Streptococcus Pneumoniae: Implications for Drug Selection. Clin. Infect. Dis. 2002, 34, 1613–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherazard, R.; Epstein, M.; Doan, T.-L.; Salim, T.; Bharti, S.; Smith, M.A. Antimicrobial Resistant Streptococcus Pneumoniae: Prevalence, Mechanisms, and Clinical Implications. Am. J. Ther. 2017, 24, e361–e369. [Google Scholar] [CrossRef]
- Prats, G. Microbiología y Parasitología Médicas, 1st ed.; Editorial Médica Panamericana: Madrid, Spain, 2013; Volume 1, ISBN 978-84-9835-688-5. [Google Scholar]
- Cag, Y.; Caskurlu, H.; Fan, Y.; Cao, B.; Vahaboglu, H. Resistance Mechanisms. Ann. Transl. Med. 2016, 4, 326. [Google Scholar] [CrossRef] [Green Version]
- Oteo Iglesias, J. La Resistencia a Los Antibióticos. La Amenaza de Las Superbacterias; Los Libros de la Catarata: Madrid, Spain, 2016; ISBN 978-84-9097-214-4. [Google Scholar]
- Giráldez-Pérez, R.M.; Grueso, E.M.; Jiménez-Aguayo, R.; Carbonero, A.; González-Bravo, M.; Kuliszewska, E.; Prado-Gotor, R. Use of Nanoparticles to Prevent Resistance to Antibiotics—Synthesis and Characterization of Gold Nanosystems Based on Tetracycline. Pharmaceutics 2022, 14, 1941. [Google Scholar] [CrossRef]
- Abdullah, S.S.; Masood, S.; Zaneb, H.; Rabbani, I.; Akbar, J.; Kuthu, Z.H.; Masood, A.; Vargas-Bello-pérez, E. Effects of Copper Nanoparticles on Performance, Muscle and Bone Characteristics and Serum Metabolites in Broilers | Efeitos de Nanopartículas de Cobre No Desempenho, Características Musculares e Ósseas e Metabólitos Séricos Em Frangos de Corte. Braz. J. Biol. 2024, 84, e261578. [Google Scholar] [CrossRef]
- Al-Awsi, G.R.L.; Alameri, A.A.; Al-Dhalimy, A.M.B.; Gabr, G.A.; Kianfar, E. Application of Nano-Antibiotics in the Diagnosis and Treatment of Infectious Diseases | Aplicação de Nanoantibióticos No Diagnóstico e Tratamento de Doenças Infecciosas. Braz. J. Biol. 2023, 84, e264946. [Google Scholar] [CrossRef]
- Fan, Y.; Marioli, M.; Zhang, K. Analytical Characterization of Liposomes and Other Lipid Nanoparticles for Drug Delivery. J. Pharm. Biomed. Anal. 2021, 192, 113642. [Google Scholar] [CrossRef]
- Altun, E.; Aydogdu, M.O.; Chung, E.; Ren, G.; Homer-Vanniasinkam, S.; Edirisinghe, M. Metal-Based Nanoparticles for Combating Antibiotic Resistance. Appl. Phys. Rev. 2021, 8, 041303. [Google Scholar] [CrossRef]
- Planchon, M.; Ferrari, R.; Guyot, F.; Gélabert, A.; Menguy, N.; Chanéac, C.; Thill, A.; Benedetti, M.F.; Spalla, O. Interaction between Escherichia Coli and TiO2 Nanoparticles in Natural and Artificial Waters. Colloids Surf. B Biointerfaces 2013, 102, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kiwi, J.; Nadtochenko, V. Evidence for the Mechanism of Photocatalytic Degradation of the Bacterial Wall Membrane at the TiO2 Interface by ATR-FTIR and Laser Kinetic Spectroscopy. Langmuir 2005, 21, 4631–4641. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Cheng, S.; Singh, S. Oxidative Stress-Mediated Genotoxic Effect of Zinc Oxide Nanoparticles on Deinococcus Radiodurans. 3 Biotech 2020, 10, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, S.-H.; Hong, J.; McGuffie, M.; Yeom, B.; Vanepps, J.S.; Kotov, N.A. Shape-Dependent Biomimetic Inhibition of Enzyme by Nanoparticles and Their Antibacterial Activity. ACS Nano 2015, 9, 9097–9105. [Google Scholar] [CrossRef]
- Khan, S.T.; Malik, A.; Wahab, R.; Abd-Elkader, O.H.; Ahamed, M.; Ahmad, J.; Musarrat, J.; Siddiqui, M.A.; Al-Khedhairy, A.A. Synthesis and Characterization of Some Abundant Nanoparticles, Their Antimicrobial and Enzyme Inhibition Activity. Acta Microbiol. Immunol. Hung. 2017, 64, 203–216. [Google Scholar] [CrossRef] [Green Version]
- Quinteros, M.A.; Cano Aristizábal, V.; Dalmasso, P.R.; Paraje, M.G.; Páez, P.L. Oxidative Stress Generation of Silver Nanoparticles in Three Bacterial Genera and Its Relationship with the Antimicrobial Activity. Toxicol. Vitr. 2016, 36, 216–223. [Google Scholar] [CrossRef]
- Seong, M.; Lee, D.G. Silver Nanoparticles Against Salmonella Enterica Serotype Typhimurium: Role of Inner Membrane Dysfunction. Curr. Microbiol. 2017, 74, 661–670. [Google Scholar] [CrossRef]
- Li, R.; Chen, J.; Cesario, T.C.; Wang, X.; Yuan, J.S.; Rentzepis, P.M. Synergistic Reaction of Silver Nitrate, Silver Nanoparticles, and Methylene Blue against Bacteria. Proc. Natl. Acad. Sci. USA 2016, 113, 13612–13617. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, H.; Jiang, X. Deploying Gold Nanomaterials in Combating Multi-Drug-Resistant Bacteria. ACS Nano 2022, 16, 10066–10087. [Google Scholar] [CrossRef]
- Mohamed, M.M.; Fouad, S.A.; Elshoky, H.A.; Mohammed, G.M.; Salaheldin, T.A. Antibacterial Effect of Gold Nanoparticles against Corynebacterium Pseudotuberculosis. Int. J. Vet. Sci. Med. 2017, 5, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, P.S.; Kim, C.K.; Han, G.; Forbes, N.S.; Rotello, V.M. Efficient Gene Delivery Vectors by Tuning the Surface Charge Density of Amino Acid-Functionalized Gold Nanoparticles. ACS Nano 2008, 2, 2213–2218. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Rong, K.; Li, J.; Yang, H.; Chen, R. Size-Dependent Antibacterial Activities of Silver Nanoparticles against Oral Anaerobic Pathogenic Bacteria. J. Mater. Sci. Mater. Med. 2013, 24, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Queitsch, U.; Mohn, E.; Schäffel, F.; Schultz, L.; Rellinghaus, B.; Blüher, A.; Mertig, M. Regular Arrangement of Nanoparticles from the Gas Phase on Bacterial Surface-Protein Layers. Appl. Phys. Lett. 2007, 90, 113114. [Google Scholar] [CrossRef]
- Brycki, B.; Koziróg, A.; Kowalczyk, I.; Pospieszny, T.; Materna, P.; Marciniak, J. Synthesis, Structure, Surface and Antimicrobial Properties of New Oligomeric Quaternary Ammonium Salts with Aromatic Spacers. Molecules 2017, 22, 1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grueso, E.; Roldan, E.; Perez-Tejeda, P.; Kuliszewska, E.; Molero, B.; Brecker, L.; Giráldez-Pérez, R.M. Reversible DNA Compaction Induced by Partial Intercalation of 16-Ph-16 Gemini Surfactants: Evidence of Triple Helix Formation. Phys. Chem. Chem. Phys. 2018, 20, 24902–24914. [Google Scholar] [CrossRef]
- Nardone, L.L.; Andrews, S.B. Cell Line A549 as a Model of the Type II Pneumocyte. Phospholipid Biosynthesis from Native and Organometallic Precursors. Biochim. Biophys. Acta BBA Lipids Lipid Metab. 1979, 573, 276–295. [Google Scholar] [CrossRef] [PubMed]
- Laschewsky, A.; Lunkenheimer, K.; Rakotoaly, R.H.; Wattebled, L. Spacer Effects in Dimeric Cationic Surfactants. Colloid Polym. Sci. 2005, 283, 469–479. [Google Scholar] [CrossRef]
- Hajian, R.; Shams, N.; Azarinezhad, N. Study on the Interaction of Amoxicillin and DNA by Spectroscopic and Voltammetric Studies. Asian J. Chem. 2010, 22, 8007–8016. [Google Scholar]
- Giráldez-Pérez, R.M.; Grueso, E.; Montero-Hidalgo, A.J.; Luque, R.M.; Carnerero, J.M.; Kuliszewska, E.; Prado-Gotor, R. Gold Nanosystems Covered with Doxorubicin/DNA Complexes: A Therapeutic Target for Prostate and Liver Cancer. Int. J. Mol. Sci. 2022, 23, 15575. [Google Scholar] [CrossRef]
- Giráldez-Pérez, R.M.; Grueso, E.; Lhamyani, S.; Perez-Tejeda, P.; Gentile, A.M.; Kuliszewska, E.; Roman-Perez, J.; El Bekay, R. MiR-21/Gemini Surfactant-Capped Gold Nanoparticles as Potential Therapeutic Complexes: Synthesis, Characterization and in Vivo Nanotoxicity Probes. J. Mol. Liq. 2020, 313, 113577. [Google Scholar] [CrossRef]
- Giráldez-Pérez, R.M.; Grueso, E.; Domínguez, I.; Pastor, N.; Kuliszewska, E.; Prado-Gotor, R.; Requena-Domenech, F. Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy. Pharmaceutics 2021, 13, 423. [Google Scholar] [CrossRef]
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST Expert Rules in Antimicrobial Susceptibility Testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EUCAST. Subcommittee on MIC Distributions and ECOFFs Antimicrobial Wild Type Distributions of Microorganisms. Available online: https://mic.eucast.org/search/ (accessed on 27 April 2023).
- Pham, T.-N.; Loupias, P.; Dassonville-Klimpt, A.; Sonnet, P. Drug Delivery Systems Designed to Overcome Antimicrobial Resistance. Med. Res. Rev. 2019, 39, 2343–2396. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Palazzolo, S.; Bayda, S.; Corona, G.; Toffoli, G.; Rizzolio, F. DNA Nanotechnology for Cancer Therapeutics. Theranostics 2016, 6, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Chao, J.; Liu, H.; Su, S.; Wang, L.; Huang, W.; Fan, C. Structural DNA Nanotechnology for Intelligent Drug Delivery. Small 2014, 10, 4626–4635. [Google Scholar] [CrossRef]
- Grueso, E.; López-Pérez, G.; Castellano, M.; Prado-Gotor, R. Thermodynamic and Structural Study of Phenanthroline Derivative Ruthenium Complex/DNA Interactions: Probing Partial Intercalation and Binding Properties. J. Inorg. Biochem. 2012, 106, 1–9. [Google Scholar] [CrossRef]
- Rahban, M.; Divsalar, A.; Saboury, A.A.; Golestani, A. Nanotoxicity and Spectroscopy Studies of Silver Nanoparticle: Calf Thymus DNA and K562 as Targets. J. Phys. Chem. C 2010, 114, 5798–5803. [Google Scholar] [CrossRef]
- Patel, S.; Patel, P.; Undre, S.B.; Pandya, S.R.; Singh, M.; Bakshi, S. DNA Binding and Dispersion Activities of Titanium Dioxide Nanoparticles with UV/Vis Spectrophotometry, Fluorescence Spectroscopy and Physicochemical Analysis at Physiological Temperature. J. Mol. Liq. 2016, 213, 304–311. [Google Scholar] [CrossRef]
- Prado-Gotor, R.; Grueso, E. Noncovalent Interactions of Tiopronin-Protected Gold Nanoparticles with DNA: Two Methods to Quantify Free Energy of Binding. Sci. World J. 2014, 2014, 143645. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, O.; Chavez, B.; Shaktah, R.; Garcia, P.P.; Minehan, T. Synthesis and DNA Binding Profile of Monomeric, Dimeric, and Trimeric Derivatives of Crystal Violet. Bioorg. Chem. 2019, 83, 297–302. [Google Scholar] [CrossRef]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Nucleic Acid Structure and Recognition, 1st ed.; Oxford University Press: New York, NY, USA, 2002; ISBN 0-19-850635-X. [Google Scholar]
- Prado-Gotor, R.; Grueso, E. A Kinetic Study of the Interaction of DNA with Gold Nanoparticles: Mechanistic Aspects of the Interaction. Phys. Chem. Chem. Phys. 2011, 13, 1479–1489. [Google Scholar] [CrossRef]
- Müller, W.; Crothers, D.M. Interactions of Heteroaromatic Compounds with Nucleic Acids. Eur. J. Biochem. 1975, 54, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Monnot, M.; Mauffret, O.; Lescot, E.; Fermandjian, S. Probing Intercalation and Conformational Effects of the Anticancer Drug 2-methyl-9-hydroxyellipticinium Acetate in DNA Fragments with Circular Dichroism. Eur. J. Biochem. 1992, 204, 1035–1039. [Google Scholar] [CrossRef]
- Zuber, A.; Purdey, M.; Schartner, E.; Forbes, C.; Van Der Hoek, B.; Giles, D.; Abell, A.; Monro, T.; Ebendorff-Heidepriem, H. Detection of Gold Nanoparticles with Different Sizes Using Absorption and Fluorescence Based Method. Sens. Actuators B Chem. 2016, 227, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Freitas, C.; Müller, R.H. Effect of Light and Temperature on Zeta Potential and Physical Stability in Solid Lipid Nanoparticle (SLN®) Dispersions. Int. J. Pharm. 1998, 168, 221–229. [Google Scholar] [CrossRef]
- Lee, N.-Y.; Ko, W.-C.; Hsueh, P.-R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glassman, P.M.; Muzykantov, V.R. Pharmacokinetic and Pharmacodynamic Properties of Drug Delivery Systems. J. Pharmacol. Exp. Ther. 2019, 370, 570–580. [Google Scholar] [CrossRef]
- Laakso, R.; Kristoffersson, E.; Marvola, M. Bi-Exponential First-Order Release Kinetics of Indomethacin from Tablets Containing Polysorbate 80. Int. J. Pharm. 1984, 19, 35–42. [Google Scholar] [CrossRef]
- Yus, C.; Irusta, S.; Sebastian, V.; Arruebo, M. Controlling Particle Size and Release Kinetics in the Sustained Delivery of Oral Antibiotics Using PH-Independent Mucoadhesive Polymers. Mol. Pharm. 2020, 17, 3314–3327. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.L.; Surewaard, B.G.J. Neutrophil Evasion Strategies by Streptococcus Pneumoniae and Staphylococcus Aureus. Cell Tissue Res. 2018, 371, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Perumal, E. Metal Oxide Nanoparticles as Antimicrobial Agents: A Promise for the Future. Int. J. Antimicrob. Agents 2017, 49, 137–152. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Su, Y.-P.; Chen, C.-C.; Jia, G.; Wang, H.-L.; Wu, J.C.G.; Lin, J.-G. Relationship between Antibacterial Activity of Chitosan and Surface Characteristics of Cell Wall. Acta Pharmacol. Sin. 2004, 25, 932–936. [Google Scholar] [PubMed]
- Beveridge, T.J. Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef]
- Nel, A.; Xia, T.; Mädler, L.; Li, N. Toxic Potential of Materials at the Nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef] [Green Version]
- Padil, V.V.T.; Černík, M. Green Synthesis of Copper Oxide Nanoparticles Using Gum Karaya as a Biotemplate and Their Antibacterial Application. Int. J. Nanomed. 2013, 8, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Hajipour, M.J.; Fromm, K.M.; Akbar Ashkarran, A.; Jimenez de Aberasturi, D.; Larramendi, I.R.D.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial Properties of Nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.R.; Imlay, J.A. How Escherichia Coli Tolerates Profuse Hydrogen Peroxide Formation by a Catabolic Pathway. J. Bacteriol. 2013, 195, 4569–4579. [Google Scholar] [CrossRef] [Green Version]
- Suárez, C.; Gudiol, F. Beta-Lactam Antibiotics|Antibióticos Betalactámicos. Enfermedades Infecc. Microbiol. Clin. 2009, 27, 116–129. [Google Scholar] [CrossRef]
- Schanholtzer, C.J.; Peterson, L.R. False Susceptible Penicillin G MICs for Streptococcus Pneumoniae with a Commercial Microdilution System. Am. J. Clin. Pathol. 1986, 85, 626–629. [Google Scholar] [CrossRef]
- Li, X.; Robinson, S.M.; Gupta, A.; Saha, K.; Jiang, Z.; Moyano, D.F.; Sahar, A.; Riley, M.A.; Rotello, V.M. Functional Gold Nanoparticles as Potent Antimicrobial Agents against Multi-Drug-Resistant Bacteria. ACS Nano 2014, 8, 10682–10686. [Google Scholar] [CrossRef] [Green Version]
- Niemirowicz, K.; Swiecicka, I.; Wilczewska, A.Z.; Misztalewska, I.; Kalska-Szostko, B.; Bienias, K.; Bucki, R.; Car, H. Gold-Functionalized Magnetic Nanoparticles Restrict Growth of Pseudomonas Aeruginosa. Int. J. Nanomed. 2014, 9, 2217–2224. [Google Scholar] [CrossRef] [Green Version]
- Uma Suganya, K.S.; Govindaraju, K.; Ganesh Kumar, V.; Stalin Dhas, T.; Karthick, V.; Singaravelu, G.; Elanchezhiyan, M. Blue Green Alga Mediated Synthesis of Gold Nanoparticles and Its Antibacterial Efficacy against Gram Positive Organisms. Mater. Sci. Eng. C 2015, 47, 351–356. [Google Scholar] [CrossRef]
- Ahmad, A.; Wei, Y.; Ullah, S.; Shah, S.I.; Nasir, F.; Shah, A.; Iqbal, Z.; Tahir, K.; Khan, U.A.; Yuan, Q. Synthesis of Phytochemicals-Stabilized Gold Nanoparticles and Their Biological Activities against Bacteria and Leishmania. Microb. Pathog. 2017, 110, 304–312. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. Assay of Catalases and Peroxidases. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1955; Volume 2; pp. 764–775. ISBN 0076-6879. [Google Scholar]
- Lu, X.Y.; Wu, D.C.; Li, Z.J.; Chen, G.Q. Polymer Nanoparticles. Prog. Mol. Biol. Transl. Sci. 2011, 104, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Miao, L.; Zhong, G.; Lin, C.-H.; Dargazangy, R.; Alexander-Katz, A. Understanding the Synergistic Effect of Physicochemical Properties of Nanoparticles and Their Cellular Entry Pathways. Commun. Biol. 2020, 3, 205. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Beyth, N.; Houri-Haddad, Y.; Domb, A.; Khan, W.; Hazan, R. Alternative Antimicrobial Approach: Nano-Antimicrobial Materials. Evid. Based Complement. Altern. Med. 2015, 2015, 246012. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The Antimicrobial Activity of Nanoparticles: Present Situation and Prospects for the Future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deplanche, K.; Caldelari, I.; Mikheenko, I.P.; Sargent, F.; Macaskie, L.E. Involvement of Hydrogenases in the Formation of Highly Catalytic Pd(0) Nanoparticles by Bioreduction of Pd(II) Using Escherichia Coli Mutant Strains. Microbiology 2010, 156, 2630–2640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kean, T.; Thanou, M. Biodegradation, Biodistribution and Toxicity of Chitosan. Adv. Drug Deliv. Rev. 2010, 62, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lotfipour, F.; Shahi, S.; Farjami, A.; Salatin, S.; Mahmoudian, M.; Dizaj, S.M. Safety and Toxicity Issues of Therapeutically Used Nanoparticles from the Oral Route. Biomed. Res. Int. 2021, 2021, 9322282. [Google Scholar] [CrossRef]
- Rajchakit, U.; Sarojini, V. Recent Developments in Antimicrobial-Peptide-Conjugated Gold Nanoparticles. Bioconjug. Chem. 2017, 28, 2673–2686. [Google Scholar] [CrossRef]
- Elbehiry, A.; Al-Dubaib, M.; Marzouk, E.; Moussa, I. Antibacterial Effects and Resistance Induction of Silver and Gold Nanoparticles against Staphylococcus Aureus-Induced Mastitis and the Potential Toxicity in Rats. Microbiologyopen 2019, 8, e00698. [Google Scholar] [CrossRef] [Green Version]
- Elsaesser, A.; Howard, C.V. Toxicology of Nanoparticles. Adv. Drug Deliv. Rev. 2012, 64, 129–137. [Google Scholar] [CrossRef]
















| Sample | Sample Composition | Zeta Potential (mV) | Size (nm); Population % |
|---|---|---|---|
| 1 | Au@16-mph-16 | 67.8 ± 2.6 | (2.6 ± 0.3) |
| 2 | DNA/AMOX CDNA = 6.8 × 10−6 M/CAMOX = 6.8 × 10−6 M | −73.9 ± 1.1 | d1 = (85 ± 16.4); 6.8% d2 = (416 ± 42.9); 93.2% |
| 3 | NS1 (CAu@16-mph-16 = 3.4 nM) | −36.7 ± 1.0 | d1 = (44 ± 8); 95.5% d2 = (205 ± 52); 4.5% |
| 4 | NS2 (CAu@16-mph-16 = 32.6 nM ) | −45.1± 1.1 | (69.0 ± 1.0) |
| Formulation | Equation | Lag. Time (s) | Correlation Coefficient (R2) |
|---|---|---|---|
| NS1 | 0 | 0.977 | |
| NS2 | 0 | 0.920 |
| E. coli | S. aureus | S. pneumoniae | |
|---|---|---|---|
| NP1 | - | - | 0.53 |
| NP2 | 68 | - | 0.53 |
| AMOX | 34 | - | 0.53 |
| Mueller Hinton media | - | - | 0.53 |
| NS1 | 17 | - | 0.53 |
| NS2 | 17 | - | 0.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giráldez-Pérez, R.M.; Grueso, E.M.; Carbonero, A.; Álvarez Márquez, J.; Gordillo, M.; Kuliszewska, E.; Prado-Gotor, R. Synergistic Antibacterial Effects of Amoxicillin and Gold Nanoparticles: A Therapeutic Option to Combat Antibiotic Resistance. Antibiotics 2023, 12, 1275. https://doi.org/10.3390/antibiotics12081275
Giráldez-Pérez RM, Grueso EM, Carbonero A, Álvarez Márquez J, Gordillo M, Kuliszewska E, Prado-Gotor R. Synergistic Antibacterial Effects of Amoxicillin and Gold Nanoparticles: A Therapeutic Option to Combat Antibiotic Resistance. Antibiotics. 2023; 12(8):1275. https://doi.org/10.3390/antibiotics12081275
Chicago/Turabian StyleGiráldez-Pérez, Rosa M., Elia M. Grueso, Alfonso Carbonero, Juan Álvarez Márquez, Mirian Gordillo, Edyta Kuliszewska, and Rafael Prado-Gotor. 2023. "Synergistic Antibacterial Effects of Amoxicillin and Gold Nanoparticles: A Therapeutic Option to Combat Antibiotic Resistance" Antibiotics 12, no. 8: 1275. https://doi.org/10.3390/antibiotics12081275