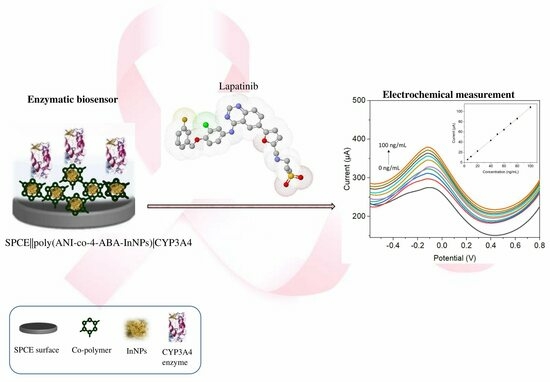
Novel Cytochrome P450-3A4 Enzymatic Nanobiosensor for Lapatinib (a Breast Cancer Drug) Developed on a Poly(anilino-co-4-aminobenzoic Acid-Green-Synthesised Indium Nanoparticle) Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Material Characterization Instrumentation
2.3. Electrochemical Measurements
2.4. Green Synthesis of Indium Nanoparticles
2.5. Synthesis of Poly(anilino-co-4-aminobenzoic Acid-Indium Nanoparticles) (Poly(ANI-co-4-ABA-InNPs)) Nanocomposites
2.6. Preparation of SPCE||Poly(ANI-co-4-ABA-InNPs)|CYP3A4|PEG Nanobiosensor
3. Results and Discussions
3.1. Optical and Structural Properties of Poly(ANI-co-4-ABA-InNPs) Nanocomposite
FTIR and UV-VIS
3.2. Morphological Properties of Poly(ANI-co-4-ABA-InNPs) Nanocomposite
3.2.1. TEM and XRD
3.2.2. SEM and Energy-Dispersive X-ray (EDX)
3.3. Electrochemical Evaluation of Poly(ANI-co-4-ABA-InNPs) Prepared as Thin Films onto SPCEs
3.3.1. Electrochemical Characterisation of Nanosensor
3.3.2. Optimization Studies
3.3.3. Electrochemistry of the Step-by-Step Development of Nanobiosensor
3.4. Electrochemical Detection of Lapatinib Using the SPCE||Poly(ANI-co-4-ABA-InNPs)|CYP3A4|PEG Nanobiosensor
3.4.1. Mechanism for the Electrochemical Detection of LAPA
3.4.2. SPCE||Poly(ANI-co-4-ABA-InNPs)|CYP3A4|PEG Nanobiosensor for the Detection of LAPA
3.4.3. Control Studies of SPCE||Poly(ANI-co-4-ABA-InNPs) Responses to Lapatinib
3.5. Selectivity, Stability, and Reproducibility of Lapatinib Biosensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. Global Cancer Observatory: Cancer Today. Lyon Fr. Int. Agency Res. Cancer 2018, 3, 2019. [Google Scholar]
- Dowlatshahi, K.; Francescatti, D.S.; Bloom, K.J. Scientific Paper Laser Therapy for Small Breast Cancers. Am. J. Surg. 2002, 184, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.F.; De Almeida, D.R.Q.; Terra, L.F.; Baptista, M.S.; Labriola, L. Photodynamic Therapy in Cancer Treatment—An Update Review. J. Cancer Metastasis Treat. 2019, 5, 1–32. [Google Scholar] [CrossRef]
- Bhadra, P.; Deb, A. Targeted Therapy for Cancer in Women Nutraceuticals View Project Role of Phytochemicals in Human Physiological Disorders: Diabetes and Obesity View Project. Indian J. Nat. Sci. 2020, 10, 20609–20616. [Google Scholar]
- Drǎgǎnescu, M.; Carmocan, C. Hormone Therapy in Breast Cancer. Chir. Rom. 2017, 112, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Trapani, J.A.; Darcy, P.K. Immunotherapy of Cancer. Aust. Fam. Physician 2017, 46, 194–198. [Google Scholar]
- Oh, D.Y.; Bang, Y.J. HER2-Targeted Therapies—A Role beyond Breast Cancer. Nat. Rev. Clin. Oncol. 2020, 17, 33–48. [Google Scholar] [CrossRef]
- Agelaki, S.; Kalykaki, A.; Markomanolaki, H.; Papadaki, M.A.; Kallergi, G.; Hatzidaki, D.; Kalbakis, K.; Mavroudis, D.; Georgoulias, V. Efficacy of Lapatinib in Therapy-Resistant HER2-Positive Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0123683. [Google Scholar] [CrossRef]
- Ates, H.C.; Roberts, J.A.; Lipman, J.; Cass, A.E.G.; Urban, G.A.; Dincer, C. On-Site Therapeutic Drug Monitoring. Trends Biotechnol. 2020, 38, 1262–1277. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 116. [Google Scholar] [CrossRef]
- Sanjabi, S.; Lear, S. New Cytometry Tools for Immune Monitoring during Cancer Immunotherapy. Cytom. B Clin. Cytom. 2021, 100, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Perfézou, M.; Turner, A.; Merkoçi, A. Cancer Detection Using Nanoparticle-Based Sensors. Chem. Soc. Rev. 2012, 41, 2606–2622. [Google Scholar] [CrossRef]
- Arndt, N.; Tran, H.D.N.; Zhang, R.; Xu, Z.P.; Ta, H.T. Different Approaches to Develop Nanosensors for Diagnosis of Diseases. Adv. Sci. 2020, 7, 2001476. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Franke, C.; Ajayi, R.F.; Uhuo, O.; Januarie, K.; Iwuoha, E. Metallodendrimer-Sensitised Cytochrome P450 3A4 Electrochemical Biosensor for TB Drugs. Electroanalysis 2020, 32, 3075–3085. [Google Scholar] [CrossRef]
- Oranzie, M.; Douman, S.; Uhuo, O.; Mokwebo, K.; Sanga, N.; Iwuoha, E. Chronocoulometric Signalling of BNP Using a Novel Quantum Dot Aptasensor. Analyst 2022, 147, 4829–4837. [Google Scholar] [CrossRef]
- Mokwebo, K.V.; Oluwafemi, O.S.; Arotiba, O.A. An Electrochemical Cholesterol Biosensor Based on a CdTe/CdSe/ZnSe Quantum Dots—Poly (Propylene Imine) Dendrimer Nanocomposite Immobilisation Layer. Sensors 2018, 18, 3368. [Google Scholar] [CrossRef]
- Sunday, C.E.; Masikini, M.; Wilson, L.; Rassie, C.; Waryo, T.; Baker, P.G.L.; Iwuoha, E.I. Application on Gold Nanoparticles-Dotted 4-Nitrophenylazo Graphene in a Label-Free Impedimetric Deoxynivalenol Immunosensor. Sensors 2015, 15, 3854–3871. [Google Scholar] [CrossRef]
- Masikini, M.; Williams, A.R.; Sunday, C.E.; Waryo, T.T.; Nxusani, E.; Wilson, L.; Qakala, S.; Bilibana, M.; Douman, S.; Jonnas, A.; et al. Label Free Poly(2,5-Dimethoxyaniline)-Multi-Walled Carbon Nanotubes Impedimetric Immunosensor for Fumonisin B1 Detection. Materials 2016, 9, 273. [Google Scholar] [CrossRef]
- Tefera, M.; Tessema, M.; Admassie, S.; Ward, M.; Phelane, L.; Iwuoha, E.I.; Baker, P.G.L. Electrochemical Application of Cobalt Nanoparticles-Polypyrrole Composite Modified Electrode for the Determination of Phoxim. Anal. Chim. Acta X 2021, 9, 100077. [Google Scholar] [CrossRef]
- Jijana, A.N.; Feleni, U.; Ndangili, P.M.; Bilibana, M.; Ajayi, R.F.; Iwuoha, E.I. Quantum Dot-Sensitised Estrogen Receptor-α-Based Biosensor for 17β-Estradiol. Biosensors 2023, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Ajay, R.F.I.; Tshoko, S.; Mgwili, Y.; Nqunqa, S.; Mulaudzi, T.; Mayedwa, N.; Iwuoha, E. Green Method Synthesised Graphene-Silver Electrochemical Nanobiosensors for Ethambutol and Pyrazinamide. Processes 2020, 8, 879. [Google Scholar] [CrossRef]
- Dhand, V.; Soumya, L.; Bharadwaj, S.; Chakra, S.; Bhatt, D.; Sreedhar, B. Green Synthesis of Silver Nanoparticles Using Coffea Arabica Seed Extract and Its Antibacterial Activity. Mater. Sci. Eng. C 2016, 58, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Tetteh, E.K.; Rathilal, S. Application of Magnetized Nanomaterial for Textile Effluent Remediation Using Response Surface Methodology. Mater. Today Proc. 2021, 38, 700–711. [Google Scholar] [CrossRef]
- Masoudi Asil, S.; Ahlawat, J.; Guillama Barroso, G.; Narayan, M. Nanomaterial Based Drug Delivery Systems for the Treatment of Neurodegenerative Diseases. Biomater. Sci. 2020, 8, 4088–4107. [Google Scholar] [CrossRef] [PubMed]
- Fytianos, G.; Rahdar, A.; Kyzas, G.Z. Nanomaterials in Cosmetics: Recent Updates. Nanomaterials 2020, 10, 979. [Google Scholar] [CrossRef]
- Adhikari, S.; Adhikari, U.; Mishra, A.; Guragain, B.S. Nanomaterials for Diagnostic, Treatment and Prevention of COVID-19. Appl. Sci. Technol. Ann. 2020, 1, 155–164. [Google Scholar] [CrossRef]
- Dong, Y.Z.; Choi, K.; Kwon, S.H.; Nam, J.-D.; Choi, H.J. Nanoparticles Functionalized by Conducting Polymers and Their Electrorheological and Magnetorheological Applications. Polymers 2020, 12, 204. [Google Scholar] [CrossRef]
- Kazemi, F.; Naghib, S.M.; Zare, Y.; Rhee, K.Y. Biosensing Applications of Polyaniline (PANI)-Based Nanocomposites: A Review. Polym. Rev. 2021, 61, 553–597. [Google Scholar] [CrossRef]
- da Silva, W.; Ghica, M.E.; Ajayi, R.F.; Iwuoha, E.I.; Brett, C.M.A. Impedimetric Sensor for Tyramine Based on Gold Nanoparticle Doped-Poly(8-Anilino-1-Naphthalene Sulphonic Acid) Modified Gold Electrodes. Talanta 2019, 195, 604–612. [Google Scholar] [CrossRef]
- da Silva, W.; Ghica, M.E.; Ajayi, R.F.; Iwuoha, E.I.; Brett, C.M.A. Tyrosinase Based Amperometric Biosensor for Determination of Tyramine in Fermented Food and Beverages with Gold Nanoparticle Doped Poly(8-Anilino-1-Naphthalene Sulphonic Acid) Modified Electrode. Food Chem. 2019, 282, 18–26. [Google Scholar] [CrossRef]
- Guan, H.; Gong, D.; Song, Y.; Han, B.; Zhang, N. Biosensor Composed of Integrated Glucose Oxidase with Liposome Microreactors/Chitosan Nanocomposite for Amperometric Glucose Sensing. Colloids Surf. A Physicochem. Eng. Asp. 2019, 574, 260–267. [Google Scholar] [CrossRef]
- Muthusankar, E.; Ragupathy, D. Graphene/Poly(Aniline-Co-Diphenylamine) Nanohybrid for Ultrasensitive Electrochemical Glucose Sensor. Nano-Struct. Nano-Objects 2019, 20, 100390. [Google Scholar] [CrossRef]
- Dogan-Topal, B.; Bozal-Palabiyik, B.; Ozkan, S.A.; Uslu, B. Investigation of Anticancer Drug Lapatinib and Its Interaction with DsDNA by Electrochemical and Spectroscopic Techniques. Sens. Actuators B Chem. 2014, 194, 185–194. [Google Scholar] [CrossRef]
- Aksoz, B.; Dogan Topal, B. Effect of Triton X-100 on the Electrochemical Behavior of Hydrophobic Lapatinib Used in the Treatment of Breast Cancer: A First Electroanalytical Study. J. Electrochem. Soc. 2021, 168, 076506. [Google Scholar] [CrossRef]
- Mathad, A.S.; Seetharamappa, J.; Kalanur, S.S. β-Cyclodextrin Anchored Neem Carbon Dots for Enhanced Electrochemical Sensing Performance of an Anticancer Drug, Lapatinib via Host-Guest Inclusion. J. Mol. Liq. 2022, 350, 118582. [Google Scholar] [CrossRef]
- Kocan, G.P.; Huang, M.; Li, F.; Pai, S. A sensitive LC-MS-MS assay for the determination of lapatinib in human plasma in subjects with end-stage renal disease receiving hemodialysis. J. Chromatogr. B 2018, 1097–1098, 74–82. [Google Scholar] [CrossRef]
- Fusco, G.; Gallo, F.; Tortolini, C.; Bollella, P.; Ietto, F.; De Mico, A.; D’Annibale, A.; Antiochia, R.; Favero, G.; Mazzei, F. AuNPs-Functionalized PANABA-MWCNTs Nanocomposite-Based Impedimetric Immunosensor for 2,4-Dichlorophenoxy Acetic Acid Detection. Biosens. Bioelectron. 2017, 93, 52–56. [Google Scholar] [CrossRef]
- Yanuriati, A.; Marseno, D.W.; Rochmadi; Harmayani, E. Characteristics of Glucomannan Isolated from Fresh Tuber of Porang (Amorphophallus Muelleri Blume). Carbohydr. Polym. 2017, 156, 56–63. [Google Scholar] [CrossRef]
- Costa, L.H.; Hemmer, J.V.; Wanderlind, E.H.; Gerlach, O.M.S.; Santos, A.L.H.; Tamanaha, M.S.; Bella-Cruz, A.; Corrêa, R.; Bazani, H.A.G.; Radetski, C.M.; et al. Green Synthesis of Gold Nanoparticles Obtained from Algae Sargassum Cymosum: Optimization, Characterization and Stability. Bionanoscience 2020, 10, 1049–1062. [Google Scholar] [CrossRef]
- Li, Y.; Yang, C.X.; Qian, H.L.; Zhao, X.; Yan, X.P. Carboxyl-Functionalized Covalent Organic Frameworks for the Adsorption and Removal of Triphenylmethane Dyes. ACS Appl. Nano Mater. 2019, 2, 7290–7298. [Google Scholar] [CrossRef]
- Rahayu, I.; Eddy, D.R.; Novianty, A.R.; Rukiah; Anggreni, A.; Bahti, H.; Hidayat, S. The Effect of Hydrochloric Acid-Doped Polyaniline to Enhance the Conductivity. In Proceedings of the IOP Conference Series: Materials Science and Engineering, Semarang, Indonesia, 7–8 September 2018; Institute of Physics Publishing: Bristol, UK, 2019; Volume 509. [Google Scholar]
- Rivas, B.L.; Sánchez, C.O. Poly(2-) and (3-Aminobenzoic Acids) and Their Copolymers with Aniline: Synthesis, Characterization, and Properties. J. Appl. Polym. Sci. 2003, 89, 2641–2648. [Google Scholar] [CrossRef]
- Rostami, A.; Omrani, A.; Hamedian, N. Flower-like Morphology of Poly(m-Aminobenzoic Acid) Deposited at GC Surface in the Presence of Silica Nanoparticles: Electropolymerization and Characterization. Monatsh. Chem. 2013, 144, 1775–1782. [Google Scholar] [CrossRef]
- Shen, C.; Xu, N.; Guan, R.; Yue, L.; Zhang, W. Highly Sensitive Ethanol Gas Sensor Based on In2O3 Spheres. Ionics 2021, 27, 3647–3653. [Google Scholar] [CrossRef]
- Zare, E.N.; Lakouraj, M.M.; Ramezani, A. Efficient Sorption of Pb(II) from an Aqueous Solution Using a Poly(Aniline-Co-3-Aminobenzoic Acid)-Based Magnetic Core-Shell Nanocomposite. New J. Chem. 2016, 40, 2521–2529. [Google Scholar] [CrossRef]
- Hassan, A.F.; Salam, H.M.A.; Mohamed, F.; Abdel-Gawad, O.F. The Optimization Performance of Fibrous Sodium Alginate Co-Polymer in Direct Methanol/Ethanol Fuel Cells. J. Polym. Environ. 2023, 31, 3664–3676. [Google Scholar] [CrossRef]
- Kong, Y.; Sha, Y.; Xue, S.; Wei, Y. Poly(Aniline-Co-p-Aminobenzoic Acid): A Conducting Copolymer with Broadened Useful Ph Range and Electrochemically Controllable Ion Exchange Behavior. J. Electrochem. Soc. 2014, 161, H249–H254. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, A. Effect of Electrode Surface on the Electrochromic Properties of Electropolymerized Poly(3,4-Ethylenedioxythiophene) Thin Films. Org. Electron. 2016, 30, 67–75. [Google Scholar] [CrossRef]
- Rosbottom, I.; Roberts, K.J.; Docherty, R. The Solid State, Surface and Morphological Properties of p-Aminobenzoic Acid in Terms of the Strength and Directionality of Its Intermolecular Synthons. CrystEngComm 2015, 17, 5768–5788. [Google Scholar] [CrossRef]
- Emi Princess Prasanna, A.; Karpagavinayagam, P.; Kulandaivel, S.; Vedhi, C. Investigation on Zinc Oxide Nanoparticle Incorporated Polyaniline Nano Composites for Solar Cell Applications. Mater. Today Proc. 2022, 48, 245–252. [Google Scholar] [CrossRef]
- Bednarczyk, K.; Matysiak, W.; Tański, T.; Janeczek, H.; Schab-Balcerzak, E.; Libera, M. Effect of Polyaniline Content and Protonating Dopants on Electroconductive Composites. Sci. Rep. 2021, 11, 7487. [Google Scholar] [CrossRef] [PubMed]
- Filep, T.; Szabó, L.; Kondor, A.C.; Jakab, G.; Szalai, Z. Evaluation of the Effect of the Intrinsic Chemical Properties of Pharmaceutically Active Compounds (PhACs) on Sorption Behaviour in Soils and Goethite. Ecotoxicol. Environ. Saf. 2021, 215, 112120. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, B.C.; Sasikala, M.; Husain, J.; Pradeep, M.V.N.; Ambika Prasad, M.V.N. Synthesis, Characterization, Electrical and Sensor Behavior of PANI/In2O3 Nano Composites. Ferroelectrics 2016, 502, 159–169. [Google Scholar] [CrossRef]
- Abdallah, A.M.; Awad, R. Sm and Er Partial Alternatives of Co in Co3O4 Nanoparticles: Probing the Physical Properties. Phys. B Condens. Matter 2021, 608, 412898. [Google Scholar] [CrossRef]
- Babu, M.H.; Podder, J.; Dev, B.C.; Sharmin, M. P to N-Type Transition with Wide Blue Shift Optical Band Gap of Spray Synthesized Cd Doped CuO Thin Films for Optoelectronic Device Applications. Surf. Interfaces 2020, 19, 100459. [Google Scholar] [CrossRef]
- Granhed, E.J.; Lindman, A.; Eklöf-Österberg, C.; Karlsson, M.; Parker, S.F.; Wahnström, G. Band: Vs. Polaron: Vibrational Motion and Chemical Expansion of Hydride Ions as Signatures for the Electronic Character in Oxyhydride Barium Titanate. J. Mater. Chem. A Mater. 2019, 7, 16211–16221. [Google Scholar] [CrossRef]
- Janeoo, S.; Sharma, M.; Singh, G.; Goswamy, J. Structural and Optical Characterization of In2O3/PANI Nanocomposite Prepared by in-Situ Polymerization. In Proceedings of the AIP Conference Proceedings, Uttar Pradesh, India, 21–25 December 2015; American Institute of Physics Inc.: College Park, MD, USA, 2016; Volume 1731. [Google Scholar]
- Zhang, D.; Li, D.; Zhou, W.; Tang, H.; Zhang, H.; Yang, A.; Liang, C. The Influence of Localized States on the Optical Absorption and Carrier Transport Properties of Acylamino Hybrid Perovskites with Tunable Electronic Structures. Chin. J. Phys. 2021, 70, 240–250. [Google Scholar] [CrossRef]
- Warsi, M.F.; Bashir, B.; Zulfiqar, S.; Aadil, M.; Khalid, M.U.; Agboola, P.O.; Shakir, I.; Yousuf, M.A.; Shahid, M. Mn1-XCuxO2/Reduced Graphene Oxide Nanocomposites: Synthesis, Characterization, and Evaluation of Visible Light Mediated Catalytic Studies. Ceram. Int. 2021, 47, 5044–5053. [Google Scholar] [CrossRef]
- Dippel, A.C.; Roelsgaard, M.; Boettger, U.; Schneller, T.; Gutowski, O.; Ruett, U. Local Atomic Structure of Thin and Ultrathin Films via Rapid High-Energy X-ray Total Scattering at Grazing Incidence. IUCrJ 2019, 6, 290–298. [Google Scholar] [CrossRef]
- Murali, A.; Barve, A.; Leppert, V.J.; Risbud, S.H.; Kennedy, I.M.; Lee, H.W.H. Synthesis and Characterization of Indium Oxide Nanoparticles. Nano Lett. 2001, 1, 287–289. [Google Scholar] [CrossRef]
- Liu, D.; Lei, W.; Qin, S.; Hou, L.; Liu, Z.; Cui, Q.; Chen, Y. Large-Scale Synthesis of Hexagonal Corundum-Type In2O 3 by Ball Milling with Enhanced Lithium Storage Capabilities. J. Mater. Chem. A Mater. 2013, 1, 5274–5278. [Google Scholar] [CrossRef]
- Kwek, D.; Crivoi, A.; Duan, F. Effects of Temperature and Particle Size on the Thermal Property Measurements of Al2O3—Water Nanofluids. J. Chem. Eng. Data 2010, 55, 5690–5695. [Google Scholar] [CrossRef]
- Haghgoo, M.; Ansari, R.; Hassanzadeh-Aghdam, M.K. The Effect of Nanoparticle Conglomeration on the Overall Conductivity of Nanocomposites. Int. J. Eng. Sci. 2020, 157, 103392. [Google Scholar] [CrossRef]
- Gosens, I.; Post, J.A.; de la Fonteyne, L.J.J.; Jansen, E.H.J.M.; Geus, J.W.; Cassee, F.R.; de Jong, W.H. Impact of Agglomeration State of Nano- and Submicron Sized Gold Particles on Pulmonary Inflammation. Part. Fibre Toxicol. 2010, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.J.; Cha, S.J.; Lee, J.Y. Highly Dispersed Indium-tin-Oxide Nanoparticles Synthesized Using in-Situ Reverse Reduction Method and Their Application to Transparent Heater for Extremely High Temperature. Powder Technol. 2018, 332, 56–62. [Google Scholar] [CrossRef]
- Goh, K.W.; Johan, M.R.; Wong, Y.H. Enhanced Structural Properties of In2O3 Nanoparticles at Lower Calcination Temperature Synthesised by Co-Precipitation Method. Micro Nano Lett. 2018, 13, 270–275. [Google Scholar] [CrossRef]
- Inagaki, C.S.; Oliveira, M.M.; Bergamini, M.F.; Marcolino-Junior, L.H.; Zarbin, A.J.G. Facile Synthesis and Dopamine Sensing Application of Three Component Nanocomposite Thin Films Based on Polythiophene, Gold Nanoparticles and Carbon Nanotubes. J. Electroanal. Chem. 2019, 840, 208–217. [Google Scholar] [CrossRef]
- Wang, H.; Ma, R.; Nienhaus, K.; Nienhaus, G.U. Formation of a Monolayer Protein Corona around Polystyrene Nanoparticles and Implications for Nanoparticle Agglomeration. Small 2019, 15, e1900974. [Google Scholar] [CrossRef]
- Qin, S.; Liu, D.; Lei, W.; Chen, Y. Synthesis of an Indium Oxide Nanoparticle Embedded Graphene Three-Dimensional Architecture for Enhanced Lithium-Ion Storage. J. Mater. Chem. A Mater. 2015, 3, 18238–18243. [Google Scholar] [CrossRef]
- Vinayak, G.K.; Sridhar, B.C.; Hussain, J.; Prasad, M.V. Synthesis, Characterization and Dielectric Behaviour of Polyaniline/In2O3 Composites. J. Adv. Phys. 2016, 5, 225–229. [Google Scholar] [CrossRef]
- Raza, A.; Hryha, E. Characterization of Spatter and Sublimation in Alloy 718 during Electron Beam Melting. Materials 2021, 14, 5953. [Google Scholar] [CrossRef] [PubMed]
- Diallo, A.; Manikandan, E.; Rajendran, V.; Maaza, M. Physical & Enhanced Photocatalytic Properties of Green Synthesized SnO2 Nanoparticles via Aspalathus Linearis. J. Alloys Compd. 2016, 681, 561–570. [Google Scholar] [CrossRef]
- Giannis, K.; Kwade, A.; Finke, J.H.; Schilde, C. The Effect of Particle Shape on the Compaction of Realistic Non-Spherical Particles—A Multi-Contact DEM Study. Pharmaceutics 2023, 15, 909. [Google Scholar] [CrossRef] [PubMed]
- Souza, E.C.C.; Rey, J.F.Q.; Muccillo, E.N.S. Synthesis and Characterization of Spherical and Narrow Size Distribution Indium Oxide Nanoparticles. Appl. Surf. Sci. 2009, 255, 3779–3783. [Google Scholar] [CrossRef]
- Wang, Y.; Jing, X. Formation of Polyaniline Nanofibers: A Morphological Study. J. Phys. Chem. B 2008, 112, 1157–1162. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, G.; Wang, Z.; Huang, T.; Wang, P. Effect of Defoaming Agent on the Properties of Cement Mortars with Hydroxyethyl Methyl Cellulose through Adjusting Air Content Gradient. Cem. Concr. Compos. 2023, 139, 105024. [Google Scholar] [CrossRef]
- Saikia, K.; Bhattacharya, K.; Sen, D.; Kaushik, S.D.; Biswas, J.; Lodha, S.; Gogoi, B.; Buragohain, A.K.; Kockenberger, W.; Deb, P. Solvent Evaporation Driven Entrapment of Magnetic Nanoparticles in Mesoporous Frame for Designing a Highly Efficient MRI Contrast Probe. Appl. Surf. Sci. 2019, 464, 567–576. [Google Scholar] [CrossRef]
- Abdul Rahman, N. Preparation and Characterizations of Poly(Aniline-Co-m-Aminobenzoic Acid)/Polystyrene Composite Nanofibers and Films. Malays. J. Anal. Sci. 2016, 20, 1033–1041. [Google Scholar] [CrossRef]
- Sebastian, J.; Samuel, J.M. Prospect of electroactive poly(2-aminobenzoic acid) and poly(2-aminobenzoic acid)-nanocomposite-Fe2O3 as antibacterial agents and antioxidants. Polym. Bull. 2023, 80, 1–17. [Google Scholar] [CrossRef]
- Abubekerov, M.; Vlček, V.; Wei, J.; Miehlich, M.E.; Quan, S.M.; Meyer, K.; Neuhauser, D.; Diaconescu, P.L. Exploring Oxidation State-Dependent Selectivity in Polymerization of Cyclic Esters and Carbonates with Zinc(II) Complexes. iScience 2018, 7, 120–131. [Google Scholar] [CrossRef]
- Ghosh, T.; Kandpal, S.; Rani, C.; Bansal, L.; Tanwar, M.; Kumar, R. Multiwavelength Color Switching from Polyaniline-Viologen Bilayer: Inching toward Versatile All-Organic Flexible Electrochromic Device. Adv. Electron. Mater. 2023, 9, 2201042. [Google Scholar] [CrossRef]
- Fu, J.; An, X.; Yao, Y.; Guo, Y.; Sun, X. Electrochemical Aptasensor Based on One Step Co-Electrodeposition of Aptamer and GO-CuNPs Nanocomposite for Organophosphorus Pesticide Detection. Sens. Actuators B Chem. 2019, 287, 503–509. [Google Scholar] [CrossRef]
- Moeketse, T.N.; Baker, P.G.; Farao, A.C.; Iwuoha, E.I. Microwave-Assisted Synthesis of Schiff Base Metal–Ligand Complexes with Copper and Nickel Centres for Electrochemical In Vitro Sensing of Nitric Oxide in an Aqueous Solution. Chemosensors 2022, 10, 175. [Google Scholar] [CrossRef]
- Askari, M.B.; Salarizadeh, P.; Seifi, M.; Zadeh, M.H.R.; Di Bartolomeo, A. ZnFe2O4 Nanorods on Reduced Graphene Oxide as Advanced Supercapacitor Electrodes. J. Alloys Compd. 2021, 860, 158497. [Google Scholar] [CrossRef]
- Pournaghi-Azar, M.H.; Habibi, B. Electropolymerization of Aniline in Acid Media on the Bare and Chemically Pre-Treated Aluminum Electrodes: A Comparative Characterization of the Polyaniline Deposited Electrodes. Electrochim. Acta 2007, 52, 4222–4230. [Google Scholar] [CrossRef]
- Salavagione, H.J.; Acevedo, D.F.; Miras, M.C.; Motheo, A.J.; Barbero, C.A. Comparative Study of 2-Amino and 3-Aminobenzoic Acid Copolymerization with Aniline Synthesis and Copolymer Properties. J. Polym. Sci. A Polym. Chem. 2004, 42, 5587–5599. [Google Scholar] [CrossRef]
- Grabowska, B.; Cukrowicz, S.; Kurleto-Kozioł, Ż.; Kaczmarska, K.; Drożyński, D.; Sitarz, M.; Bobrowski, A. Studies of Poly(Acrylic Acid-Co-Maleic Acid) Sodium Salt Intercalated Montmorillonite. Arch. Foundry Eng. 2019, 19, 67–75. [Google Scholar] [CrossRef]
- El-Enany, G.; Ahmed, M.; El-Ghaffar, A.; Emam, A.; El-Mekawy, S.A.; Abd El-Ghaffar, M.A. Electrochemical Synthesis of a Self-Doped Polyaniline and Its Supercapacitive Properties; Elsevier: Amsterdam, The Netherlands, 2013; pp. 2–13. [Google Scholar]
- Okpara, E.C.; Fayemi, O.E.; Sherif, E.S.M.; Ganesh, P.S.; Swamy, B.E.K.; Ebenso, E.E. Electrochemical Evaluation of Cd2+ and Hg2+ Ions in Water Using ZnO/Cu2ONPs/PANI Modified SPCE Electrode. Sens. Biosensing Res. 2022, 35, 100476. [Google Scholar] [CrossRef]
- Netsuwan, P.; Chaisu, W.; Phanichphant, S.; Sriwichai, S. Nanocomposite Thin Film of Poly(3-Aminobenzoic Acid) and Multiwalled Carbon Nanotubes Fabricated through an Electrochemical Method. Adv. Mater. Sci. Eng. 2014, 2014, 873028. [Google Scholar] [CrossRef]
- Batool, R.; Akhtar, M.A.; Hayat, A.; Han, D.; Niu, L.; Ahmad, M.A.; Nawaz, M.H. A Nanocomposite Prepared from Magnetite Nanoparticles, Polyaniline and Carboxy-Modified Graphene Oxide for Non-Enzymatic Sensing of Glucose. Microchim. Acta 2019, 186, 267. [Google Scholar] [CrossRef]
- Randviir, E.P. A Cross Examination of Electron Transfer Rate Constants for Carbon Screen-Printed Electrodes Using Electrochemical Impedance Spectroscopy and Cyclic Voltammetry. Electrochim. Acta 2018, 286, 179–186. [Google Scholar] [CrossRef]
- Feleni, U.; Sidwaba, U.; Makelane, H.; Iwuoha, E. Core–Shell Palladium Telluride Quantum Dot-Hemethiolate Cytochrome Based Biosensor for Detecting Indinavir Drug. J. Nanosci. Nanotechnol. 2019, 19, 7974–7981. [Google Scholar] [CrossRef]
- Diaz-Diestra, D.; Beltran-Huarac, J.; Bracho-Rincon, D.P.; González-Feliciano, J.A.; González, C.I.; Weiner, B.R.; Morell, G. Biocompatible ZnS:Mn Quantum Dots for Reactive Oxygen Generation and Detection in Aqueous Media. J. Nanoparticle Res. 2015, 17, 461. [Google Scholar] [CrossRef] [PubMed]
- Białobrzeska, W.; Firganek, D.; Czerkies, M.; Lipniacki, T.; Skwarecka, M.; Dziąbowska, K.; Cebula, Z.; Malinowska, N.; Bigus, D.; Bięga, E.; et al. Electrochemical Immunosensors Based on Screen-Printed Gold and Glassy Carbon Electrodes: Comparison of Performance for Respiratory Syncytial Virus Detection. Biosensors 2020, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, R.; Hussein, M.Z.; Yusof, N.A.; Bakar, F.A. Carbon Nanotube-Quicklime Nanocomposites Prepared Using a Nickel Catalyst Supported on Calcium Oxide Derived from Carbonate Stones. Nanomaterials 2019, 9, 1239. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, P.; Rezayi, M.; Rasouli, E.; Avan, A.; Gholami, M.; Ghayour Mobarhan, M.; Karimi, E.; Alias, Y. Early-Stage Cervical Cancer Diagnosis Based on an Ultra-Sensitive Electrochemical DNA Nanobiosensor for HPV-18 Detection in Real Samples. J. Nanobiotechnol. 2020, 18, 11. [Google Scholar] [CrossRef]
- Ozaytekin, I. The Effect of Carboxylic Acid Group on Conductivity of the Aromatic Polyazomethines and Char Composites. Polym. Compos. 2014, 35, 372–380. [Google Scholar] [CrossRef]
- Müller, M.; Agarwal, N.; Kim, J. A Cytochrome P450 3A4 Biosensor Based on Generation 4.0 PAMAM Dendrimers for the Detection of Caffeine. Biosensors 2016, 6, 44. [Google Scholar] [CrossRef]
- Megarity, C.F.; Siritanaratkul, B.; Herold, R.A.; Morello, G.; Armstrong, F.A. Electron Flow between the Worlds of Marcus and Warburg. J. Chem. Phys. 2020, 153, 225101. [Google Scholar] [CrossRef]
- Ross, N.; Hendricks-Leukes, N.; Ajayi, R.F.; Baker, P.; Iwuoha, E.I. Conductive Composite Biosensor System for Electrochemical Indinavir Drug Detection. J. Chem. 2015, 2015, 630408. [Google Scholar] [CrossRef]
- Kumar, N.; He, J.; Rusling, J.F. Electrochemical Transformations Catalyzed by Cytochrome P450s and Peroxidases. Chem. Soc. Rev. 2023, 52, 5135–5171. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Bai, X.; Yang, L. Intriguing Generative Metabolism Discovery of Reactive Metabolite Nitroso of Lapatinib and Relevant Structural Modification. Int. J. Quantum Chem. 2020, 120, e26210. [Google Scholar] [CrossRef]
- Phelane, L.; Gouveia-Caridade, C.; Barsan, M.M.; Baker, P.G.L.; Brett, C.M.A.; Iwuoha, E.I. Electrochemical Determination of Tyrosine Using a Novel Tyrosinase Multi-Walled Carbon Nanotube (MWCNT) Polysulfone Modified Glassy Carbon Electrode (GCE). Anal. Lett. 2020, 53, 308–321. [Google Scholar] [CrossRef]
- Baj-Rossi, C.; de Micheli, G.; Carrara, S. Electrochemical Detection of Anti-Breast-Cancer Agents in Human Serum by Cytochrome P450-Coated Carbon Nanotubes. Sensors 2012, 12, 6520–6537. [Google Scholar] [CrossRef]
- Nakate, U.T.; Ahmad, R.; Patil, P.; Yu, Y.T.; Hahn, Y.B. Ultra-Thin NiO Nanosheets for High Performance Hydrogen Gas Sensor Device. Appl. Surf. Sci. 2020, 506, 144971. [Google Scholar] [CrossRef]
- Panahi, Y.; Motaharian, A.; Hosseini, M.R.M.; Mehrpour, O. Highly Sensitive and Selective Nano-Molecularly Imprinted Polymer Based Electrochemical Sensor for Midazolam Drug Detection in Pharmaceutical Formulation and Human Urine Samples. Sens. Actuators B Chem. 2018, 273, 1579–1586. [Google Scholar] [CrossRef]
- Burris, H.A.; Hurwitz, H.I.; Dees, E.C.; Dowlati, A.; Blackwell, K.L.; O’Neil, B.; Marcom, P.K.; Ellis, M.J.; Overmoyer, B.; Jones, S.F.; et al. Phase I Safety, Pharmacokinetics, and Clinical Activity Study of Lapatinib (GW572016), a Reversible Dual Inhibitor of Epidermal Growth Factor Receptor Tyrosine Kinases, in Heavily Pretreated Patients with Metastatic Carcinomas. J. Clin. Oncol. 2005, 23, 5305–5313. [Google Scholar] [CrossRef]
- Wang, K.P.; Zhang, Y.C.; Zhang, X.; Shen, L. Green Preparation of Chlorine-Doped Graphene and Its Application in Electrochemical Sensor for Chloramphenicol Detection. SN Appl. Sci. 2019, 1, 157. [Google Scholar] [CrossRef]
- Kumar, K.K.; Nagoji, K.E.V.; Nadh, R.V. A Validated RP-HPLC Method for the Estimation of Lapatinib in Tablet Dosage Form Using Gemcitabine Hydrochloride as an Internal standard. Indian J. Pharm. Sci. 2012, 74, 580–583. [Google Scholar] [CrossRef]
- Escudero-Ortiz, V.; José Pérez-Ruixo, J.; Valenzuela, B. Development and Validation of a High-Performance Liquid Chromatography Ultraviolet Method for Lapatinib Quantification in Human Plasma. Ther. Drug Monit. 2013, 35, 796–802. [Google Scholar] [CrossRef]
- Panda, S.S.; Bera, R.K.V.V.; Behera, A.K.; Beg, S. Chemometrics-Assisted Development of a Liquid Chromatography Method for Estimation of Lapatinib in Tablets: A Case Study on a Novel Quality Concept. Sep. Sci. Plus 2020, 3, 12–21. [Google Scholar] [CrossRef]
- Biswal, S.; Mondal, S. Analytical Method Validation Report for Assay of Lapatinib by UPLC. Pharm. Methods 2019, 10, 9–14. [Google Scholar] [CrossRef]
- Ivaturi, R.; Sastry, M.T.; Satyaveni, S. Development and validation of stability indicating hplc method for the determination of lapatinib impurities in bulk and finished formulations. Int. J. Pharm. Sci. Res. 2017, 8, 3081–3091. [Google Scholar] [CrossRef]
- Saadat, E.; Kelishady, P.D.; Ravar, F.; Kobarfard, F.; Dorkoosh, F.A. Development and Validation of Rapid Stability-Indicating RP-HPLC-DAD Method for the Quantification of Lapatinib and Mass Spectrometry Analysis of Degraded Products. J. Chromatogr. Sci. 2015, 53, 932–939. [Google Scholar] [CrossRef]
- Bai, F.; Freeman, B.B.; Fraga, C.H.; Fouladi, M.; Stewart, C.F. Determination of Lapatinib (GW572016) in Human Plasma by Liquid Chromatography Electrospray Tandem Mass Spectrometry (LC-ESI-MS/MS). J. Chromatogr. B 2006, 831, 169–175. [Google Scholar] [CrossRef]
- Ohgami, M.; Homma, M.; Suzuki, Y.; Naito, K.; Yamada, M.; Mitsuhashi, S.; Fujisawa, F.; Kojima, H.; Kaburagi, T.; Uchiumi, K.; et al. A Simple High-Performance Liquid Chromatography for Determining Lapatinib and Erlotinib in Human Plasma. Ther. Drug Monit. 2016, 38, 657–662. [Google Scholar] [CrossRef]
- Mehrabi, A.; Rahimnejad, M.; Mohammadi, M.; Pourali, M. Electrochemical Detection of Flutamide as an Anticancer Drug with Gold Nanoparticles Modified Glassy Carbon Electrode in the Presence of Prostate Cancer Cells. J. Appl. Electrochem. 2021, 51, 597–606. [Google Scholar] [CrossRef]
- Li, G.; Zhao, M.; Zhao, L. Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry for Simultaneous Determination of 12 Anti-Tumor Drugs in Human Plasma and Its Application in Therapeutic Drug Monitoring. J. Pharm. Biomed. Anal. 2021, 206, 1–14. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, J.; Liang, M.; Zheng, H.; Zhu, C.; Wang, Y. Detection of Imatinib Based on Electrochemical Sensor Constructed Using Biosynthesized Graphene-Silver Nanocomposite. Front. Chem. 2021, 9, 670074. [Google Scholar] [CrossRef]
- Gruhl, F.J.; Rapp, B.E.; Länge, K. Biosensors for diagnostic applications. Adv. Biochem. Eng. Biotechnol. 2013, 133, 115–148. [Google Scholar] [CrossRef]
- Ko, M.; Mendecki, L.; Eagleton, A.M.; Durbin, C.G.; Stolz, R.M.; Meng, Z.; Mirica, K.A. Employing Conductive Metal-Organic Frameworks for Voltammetric Detection of Neurochemicals. J. Am. Chem. Soc. 2020, 142, 11717–11733. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.H.; Irwin, W.C.; Fesser, J.; Massey, L. Interference of Ascorbic Acid with Chemical Analytes. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2005, 42, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, Q.; Cai, J.; Yang, T.; Chen, J.; Hou, X. Electrochemical Detection Mechanism of Dopamine and Uric Acid on Titanium Nitride-Reduced Graphene Oxide Composite with and without Ascorbic Acid. Sens. Actuators B Chem. 2019, 298, 126872. [Google Scholar] [CrossRef]
- Zhang, R.; Dong, K.; Wang, Z.; Miao, R.; Lu, W.; Wu, X. Nanoparticulate Drug Delivery Strategies to Address Intestinal Cytochrome P450 Cyp3A4 Metabolism towards Personalized Medicine. Pharmaceutics 2021, 13, 1261. [Google Scholar] [CrossRef] [PubMed]













| Detection Technique | LOD (ng/mL) | LOQ (ng/mL) | Reference |
|---|---|---|---|
| SPCE||poly(ANI-co-4-ABA-InNPs)|CYP3A4|PEG in DPBS | 13.212 | 40.04 | This work |
| SPCE||poly(ANI-co-4-ABA-InNPs)|CYP3A4|PEG in human serum | 18.6 | 56.26 | This work |
| Reverse-phase HPLC | 265 | 884 | [112] |
| HPLC-UV/vis | 100 | 200 | [113] |
| Validated LC (chemometric principles) | 500 | 1000 | [114] |
| Ultra-performance liquid chromatography (UPLC) | 60 | 180 | [115] |
| LC-MS -HPLC | 10 | 40 | [116] |
| RP-HPLC-DAD | 1000 | 5000 | [117] |
| LC-ESI-MS/MS | 4.2 | 15 | [118] |
| HPLC | 125 | 300 | [119] |
| Interferent | LAPA (ng/mL) | Interferent (ng/mL) | LAPA Response (µA) | Interferent Response (µA) | LAPA + Interferent Response (µA) | Level of Interference (%) |
|---|---|---|---|---|---|---|
| Ascorbic acid | 5 | 50 | 201 | 17 | 195 | 1.013 |
| Dopamine | 5 | 50 | 119 | 24 | 109 | 1.09 |
| Uric acid | 5 | 50 | 81 | 4 | 65 | 0.8 |
| Glucose | 5 | 50 | 132 | 6 | 122 | 1.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
January, J.L.; Tshobeni, Z.Z.; Ngema, N.P.P.; Jijana, A.N.; Iwuoha, E.I.; Mulaudzi, T.; Douman, S.F.; Ajayi, R.F. Novel Cytochrome P450-3A4 Enzymatic Nanobiosensor for Lapatinib (a Breast Cancer Drug) Developed on a Poly(anilino-co-4-aminobenzoic Acid-Green-Synthesised Indium Nanoparticle) Platform. Biosensors 2023, 13, 897. https://doi.org/10.3390/bios13090897
January JL, Tshobeni ZZ, Ngema NPP, Jijana AN, Iwuoha EI, Mulaudzi T, Douman SF, Ajayi RF. Novel Cytochrome P450-3A4 Enzymatic Nanobiosensor for Lapatinib (a Breast Cancer Drug) Developed on a Poly(anilino-co-4-aminobenzoic Acid-Green-Synthesised Indium Nanoparticle) Platform. Biosensors. 2023; 13(9):897. https://doi.org/10.3390/bios13090897
Chicago/Turabian StyleJanuary, Jaymi Leigh, Ziyanda Zamaswazi Tshobeni, Nokwanda Precious Pearl Ngema, Abongile Nwabisa Jijana, Emmanuel Iheanyichukwu Iwuoha, Takalani Mulaudzi, Samantha Fiona Douman, and Rachel Fanelwa Ajayi. 2023. "Novel Cytochrome P450-3A4 Enzymatic Nanobiosensor for Lapatinib (a Breast Cancer Drug) Developed on a Poly(anilino-co-4-aminobenzoic Acid-Green-Synthesised Indium Nanoparticle) Platform" Biosensors 13, no. 9: 897. https://doi.org/10.3390/bios13090897