On-Chip Single-Cell Bioelectrical Analysis for Identification of Cell Electrical Phenotyping in Response to Sequential Electric Signal Modulation
Abstract
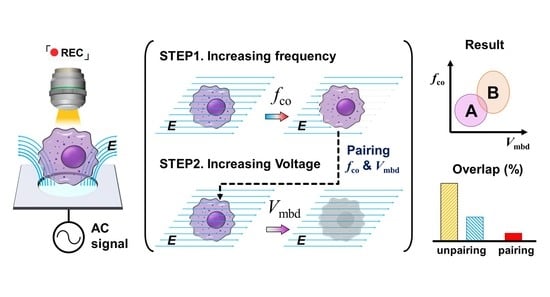
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Chip Preparation
2.3. Cell Preparation
2.4. Drug Treatment
2.5. Experimental Setup
2.6. Electric signal Configuration
2.7. Determination of DEP Crossover Frequency (fco)
2.8. Determination of Membrane Breakdown Voltage (Vmbd)
2.9. Statistical Analysis
3. Results and Discussion
3.1. Platform Concept and Working Design
3.2. Different Breast Cancer Cell Lines Were Distinguished Using the Developed Platform
3.3. Drug-Treated Breast Cancer Cells Were Distinguished Using the Developed Platform
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henslee, E.A.; Crosby, P.; Kitcatt, S.J.; Parry, J.S.W.; Bernardini, A.; Abdallat, R.G.; Braun, G.; Fatoyinbo, H.O.; Harrison, E.J.; Edgar, R.S.; et al. Rhythmic potassium transport regulates the circadian clock in human red blood cells. Nat. Commun. 2017, 8, 1978. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.P.; Kruchek, E.J.; Beale, A.D.; Kitcatt, S.J.; Qureshi, S.; Trott, Z.P.; Charbonnel, O.; Agbaje, P.A.; Henslee, E.A.; Dorey, R.A.; et al. Vm-related extracellular potentials observed in red blood cells. Sci. Rep. 2021, 11, 19446. [Google Scholar] [CrossRef]
- Pohl, E.E.; Peterson, U.; Sun, J.; Pohl, P. Changes of Intrinsic Membrane Potentials Induced by Flip-Flop of Long-Chain Fatty Acids. Biochemistry 2000, 39, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Rituper, B.; Flašker, A.; Guček, A.; Chowdhury, H.H.; Zorec, R. Cholesterol and regulated exocytosis: A requirement for unitary exocytotic events. Cell Calcium 2012, 52, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Hoettges, K.F.; Henslee, E.A.; Torcal Serrano, R.M.; Jabr, R.I.; Abdallat, R.G.; Beale, A.D.; Waheed, A.; Camelliti, P.; Fry, C.H.; van der Veen, D.R.; et al. Ten–Second Electrophysiology: Evaluation of the 3DEP Platform for high-speed, high-accuracy cell analysis. Sci. Rep. 2019, 9, 19153. [Google Scholar] [CrossRef] [Green Version]
- Binder, A.R.; Spiess, A.-N.; Pfaffl, M.W. Modelling and Differential Quantification of Electric Cell-Substrate Impedance Sensing Growth Curves. Sensors 2021, 21, 5286. [Google Scholar] [CrossRef]
- Valero, A.; Braschler, T.; Rauch, A.; Demierre, N.; Barral, Y.; Renaud, P. Tracking and synchronization of the yeast cell cycle using dielectrophoretic opacity. Lab Chip 2011, 11, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Mulhall, H.J.; Cardnell, A.; Hoettges, K.F.; Labeed, F.H.; Hughes, M.P. Apoptosis progression studied using parallel dielectrophoresis electrophysiological analysis and flow cytometry. Integr. Biol. 2015, 7, 1396–1401. [Google Scholar] [CrossRef]
- Salimi, E.; Braasch, K.; Fazelkhah, A.; Afshar, S.; Saboktakin Rizi, B.; Mohammad, K.; Butler, M.; Bridges, G.E.; Thomson, D.J. Single cell dielectrophoresis study of apoptosis progression induced by controlled starvation. Bioelectrochemistry 2018, 124, 73–79. [Google Scholar] [CrossRef]
- Taruvai Kalyana Kumar, R.; Liu, S.; Minna, J.D.; Prasad, S. Monitoring drug induced apoptosis and treatment sensitivity in non-small cell lung carcinoma using dielectrophoresis. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1877–1883. [Google Scholar] [CrossRef]
- Ahmad, M.A.; Natour, Z.A.; Mustafa, F.; Rizvi, T.A. Electrical Characterization of Normal and Cancer Cells. IEEE Access 2018, 6, 25979–25986. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, M.; Chen, D.; Zhao, X.; Xue, C.; Hao, R.; Yue, W.; Wang, J.; Chen, J. Single-Cell Electrical Phenotyping Enabling the Classification of Mouse Tumor Samples. Sci. Rep. 2016, 6, 19487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labeed, F.H.; Lu, J.; Mulhall, H.J.; Marchenko, S.A.; Hoettges, K.F.; Estrada, L.C.; Lee, A.P.; Hughes, M.P.; Flanagan, L.A. Biophysical Characteristics Reveal Neural Stem Cell Differentiation Potential. PLoS ONE 2011, 6, e25458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, T.N.G.; Jiang, A.Y.L.; Mendoza, N.S.; Ro, C.C.; Lee, D.-H.; Lee, A.P.; Flanagan, L.A. Label-free enrichment of fate-biased human neural stem and progenitor cells. Biosens. Bioelectron. 2020, 152, 111982. [Google Scholar] [CrossRef]
- Yang, J.; Huang, Y.; Wang, X.; Wang, X.-B.; Becker, F.F.; Gascoyne, P.R.C. Dielectric Properties of Human Leukocyte Subpopulations Determined by Electrorotation as a Cell Separation Criterion. Biophys. J. 1999, 76, 3307–3314. [Google Scholar] [CrossRef] [Green Version]
- Gascoyne, P.R.C.; Shim, S. Isolation of Circulating Tumor Cells by Dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef] [Green Version]
- Adekanmbi, E.O.; Srivastava, S.K. Dielectric characterization of bioparticles via electrokinetics: The past, present, and the future. Appl. Phys. Rev. 2019, 6, 041313. [Google Scholar] [CrossRef]
- Liang, W.; Zhang, K.; Yang, X.; Liu, L.; Yu, H.; Zhang, W. Distinctive translational and self-rotational motion of lymphoma cells in an optically induced non-rotational alternating current electric field. Biomicrofluidics 2015, 9, 014121. [Google Scholar] [CrossRef] [Green Version]
- Chiu, T.-K.; Zhao, Y.; Chen, D.; Hsieh, C.-H.; Wang, K.; Chou, W.-P.; Liao, C.-J.; Wang, H.-Y.; Fan, B.; Wang, J.; et al. A low-sample-loss microfluidic system for the quantification of size-independent cellular electrical property—Its demonstration for the identification and characterization of circulating tumour cells (CTCs). Sens. Actuators B-Chem. 2017, 246, 29–37. [Google Scholar] [CrossRef]
- Park, I.; Lim, J.W.; Kim, S.H.; Choi, S.; Ko, K.H.; Son, M.G.; Chang, W.-J.; Yoon, Y.R.; Yang, S.; Key, J.; et al. Variable Membrane Dielectric Polarization Characteristic in Individual Live Cells. J. Phys. Chem. Lett. 2020, 11, 7197–7203. [Google Scholar] [CrossRef]
- Bandmann, V.; Mirsanaye, A.S.; Schäfer, J.; Thiel, G.; Holstein, T.; Mikosch-Wersching, M. Membrane capacitance recordings resolve dynamics and complexity of receptor-mediated endocytosis in Wnt signalling. Sci. Rep. 2019, 9, 12999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rituper, B.; Guček, A.; Jorgačevski, J.; Flašker, A.; Kreft, M.; Zorec, R. High-resolution membrane capacitance measurements for the study of exocytosis and endocytosis. Nat. Protoc. 2013, 8, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- El-Beyrouthy, J.; Freeman, E. Characterizing the Structure and Interactions of Model Lipid Membranes Using Electrophysiology. Membranes 2021, 11, 319. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Anand, R.K. Cellular dielectrophoresis coupled with single-cell analysis. Anal. Bioanal. Chem. 2018, 410, 2499–2515. [Google Scholar] [CrossRef] [PubMed]
- Turcan, I.; Olariu, M.A. Dielectrophoretic Manipulation of Cancer Cells and Their Electrical Characterization. ACS Comb. Sci. 2020, 22, 554–578. [Google Scholar] [CrossRef] [PubMed]
- Giduthuri, A.T.; Theodossiou, S.K.; Schiele, N.R.; Srivastava, S.K. Dielectrophoresis as a tool for electrophysiological characterization of stem cells. Biophys. Rev. 2020, 1, 011304. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S.; Noshari, J.; Becker, F.F.; Stemke-Hale, K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. Electrophoresis 2013, 34, 1042–1050. [Google Scholar] [CrossRef]
- Henslee, E.A. Review: Dielectrophoresis in cell characterization. Electrophoresis 2020, 41, 1915–1930. [Google Scholar] [CrossRef]
- Brosseau, C.; Sabri, E. Resistor–capacitor modeling of the cell membrane: A multiphysics analysis. J. Appl. Phys. 2021, 129, 011101. [Google Scholar] [CrossRef]
- Honrado, C.; Bisegna, P.; Swami, N.; Caselli, F. Single-cell microfluidic impedance cytometry: From raw signals to cell phenotypes using data analytics. Lab Chip 2021, 21, 22–54. [Google Scholar] [CrossRef]
- Heileman, K.; Daoud, J.; Tabrizian, M. Dielectric spectroscopy as a viable biosensing tool for cell and tissue characterization and analysis. Biosens. Bioelectron. 2013, 49, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Rems, L.; Miklavčič, D. Tutorial: Electroporation of cells in complex materials and tissue. J. Appl. Phys. 2016, 119, 201101. [Google Scholar] [CrossRef] [Green Version]
- Kramar, P.; Miklavcic, D.; Lebar, A.M. Determination of the lipid bilayer breakdown voltage by means of linear rising signal. Bioelectrochemistry 2007, 70, 23–27. [Google Scholar] [CrossRef]
- Velikonja, A.; Kramar, P.; Miklavčič, D.; Maček Lebar, A. Specific electrical capacitance and voltage breakdown as a function of temperature for different planar lipid bilayers. Bioelectrochemistry 2016, 112, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Kramar, P.; Miklavčič, D. Effect of the cholesterol on electroporation of planar lipid bilayer. Bioelectrochemistry 2022, 144, 108004. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, H.; Lee, S.; Park, I.; Kim, Y.S.; Key, J.; Lee, S.Y.; Yang, S.; Lee, S.W. A novel automatic segmentation and tracking method to measure cellular dielectrophoretic mobility from individual cell trajectories for high throughput assay. Comput. Meth. Programs Biomed. 2020, 195, 105662. [Google Scholar] [CrossRef]
- Choi, S.; Ko, K.; Lim, J.; Kim, S.; Woo, S.-H.; Kim, Y.; Key, J.; Lee, S.; Park, I.; Lee, S. Non-Linear Cellular Dielectrophoretic Behavior Characterization Using Dielectrophoretic Tweezers-Based Force Spectroscopy inside a Microfluidic Device. Sensors 2018, 18, 3543. [Google Scholar] [CrossRef] [Green Version]
- Park, I.S.; Lee, J.; Lee, G.; Nam, K.; Lee, T.; Chang, W.-J.; Kim, H.; Lee, S.-Y.; Seo, J.; Yoon, D.S.; et al. Real-Time Analysis of Cellular Response to Small-Molecule Drugs within a Microfluidic Dielectrophoresis Device. Anal. Chem. 2015, 87, 5914–5920. [Google Scholar] [CrossRef]
- Jung, Y.-J.; Lee, T.; Choi, S.; Lee, S.-Y.; Key, J.; Yoo, Y.-M.; Chang, W.-J.; Kim, H.-S.; Lee, S.W. Selective position of individual cells without lysis on a circular window array using dielectrophoresis in a microfluidic device. Microfluid. Nanofluid. 2017, 21, 150. [Google Scholar] [CrossRef]
- Han, A.; Yang, L.; Frazier, A.B. Quantification of the Heterogeneity in Breast Cancer Cell Lines Using Whole-Cell Impedance Spectroscopy. Clin. Cancer Res. 2007, 13, 139–143. [Google Scholar] [CrossRef]
- Huang, C.; Liu, C.; Minne, B.; Ramirez Hernandez, J.E.; Stakenborg, T.; Lagae, L. Dielectrophoretic discrimination of cancer cells on a microchip. Appl. Phys. Lett. 2014, 105, 143702. [Google Scholar] [CrossRef]
- Qiao, G.; Duan, W.; Chatwin, C.; Sinclair, A.; Wang, W. Electrical properties of breast cancer cells from impedance measurement of cell suspensions. J. Phys. Conf. Ser. 2010, 224, 012081. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, S.; Park, I.; Lee, S.H.; Yeo, K.I.; Min, G.; Woo, S.-H.; Kim, Y.S.; Lee, S.Y.; Lee, S.W. On-Chip Single-Cell Bioelectrical Analysis for Identification of Cell Electrical Phenotyping in Response to Sequential Electric Signal Modulation. Biosensors 2022, 12, 1037. https://doi.org/10.3390/bios12111037
Choi S, Park I, Lee SH, Yeo KI, Min G, Woo S-H, Kim YS, Lee SY, Lee SW. On-Chip Single-Cell Bioelectrical Analysis for Identification of Cell Electrical Phenotyping in Response to Sequential Electric Signal Modulation. Biosensors. 2022; 12(11):1037. https://doi.org/10.3390/bios12111037
Chicago/Turabian StyleChoi, Seungyeop, Insu Park, Sang Hyun Lee, Kang In Yeo, Gyeongjun Min, Sung-Hun Woo, Yoon Suk Kim, Sei Young Lee, and Sang Woo Lee. 2022. "On-Chip Single-Cell Bioelectrical Analysis for Identification of Cell Electrical Phenotyping in Response to Sequential Electric Signal Modulation" Biosensors 12, no. 11: 1037. https://doi.org/10.3390/bios12111037