Allelopathic Inhibition by the Bacteria Bacillus cereus BE23 on Growth and Photosynthesis of the Macroalga Ulva prolifera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algal Culture and Identification
2.2. Experiment 1: Bacteria-Derived Allelopathic Inhibition on U. prolifera
2.2.1. Preparation of Cell-Free Filtrate from Bacillus cereus
2.2.2. Preparation of the Exposure Treatment
2.2.3. Growth
2.2.4. The Antioxidant Defense System
2.2.5. Photosynthesis System
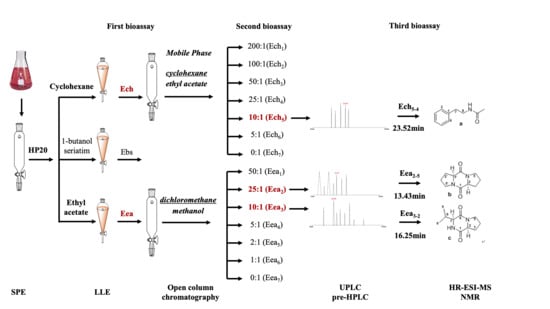
2.3. Experiment 2: Isolation and Identification the Potential Allelopathic Compounds from Cell-Free Filtrate of Bacillus cereus BE23
2.3.1. Step 1: Solid Phase and Liquid Phase Extraction of Potential Allelopathic Compounds
2.3.2. Step 2: Open Column Chromatography to Select the Potential Allelopathic Compounds
2.3.3. Step 3: Ultra- and High-Performance Liquid Chromatography to Select the Potential Allelopathic Compounds
2.3.4. Structure Identification
2.4. Statistical Analysis
3. Results
3.1. Identification of Macroalga and Bacteria
3.2. Inhibition on the Growth of U. prolifera
3.3. Response of Antioxidant System of U. prolifera
3.4. Response of PSII System of U. prolifera
3.5. Identification of Allelochemicals from Bacillus cereus BE23 Filtrate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Wang, J.T.; Xue, Q.N.; Tan, L.J.; Cai, J.; Wang, H.Y. Preliminary analysis of allelochemicals produced by the diatom Phaeodactylum tricornutum. Chemosphere 2016, 165, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.M.; Hilt, S.; Lombardo, P.; Mulderij, G. Searching for allelopathic effects of submerged macrophytes on phytoplankton—State of the art and open questions. Hydrobiologia 2007, 584, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.W.; Wang, J.T.; Tan, L.J. Characterization of allelochemicals of the diatom Chaetoceros curvisetus and the effects on the growth of Skeletonema costatum. Sci. Total Environ. 2019, 660, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Peng, Y.; Zhang, S.; Cai, G.; Li, Y.; Yang, X.; Yang, K.; Chen, Z.; Zhang, J.; Wang, H.; et al. Algicidal effects of prodigiosin on the harmful algae Phaeocystis globosa. Front. Microbiol. 2016, 7, 602. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yin, H.; Tang, S.Y.; Peng, H.; Yin, D.G.; Yang, Y.X.; Liu, Z.H.; Ding, Z. Physiological responses of Microcystis aeruginosa against the algicidal bacterium Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 2016, 127, 214–221. [Google Scholar] [CrossRef]
- Zhang, F.X.; Ye, Q.; Chen, Q.L.; Yang, K.; Zhang, D.Y.; Chen, Z.R.; Lu, S.S.; Shao, X.P.; Fan, X.Y.; Yao, L.M.; et al. Algicidal Activity of novel marine bacterium Paracoccus sp. Strain Y42 against a harmful algal-bloom-causing dinoflagellate, Prorocentrum donghaiense. Appl. Environ Microbiol. 2018, 84. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.F.; Xu, J.H.; Lu, T.; Zhang, Q.; Qu, Q.; Yang, Z.P.; Pan, X.L. Responses of unicellular alga Chlorella pyrenoidosa to allelochemical linoleic acid. Sci. Total Environ. 2018, 625, 1415–1422. [Google Scholar] [CrossRef]
- Zhao, W.; Zheng, Z.; Zhang, J.L.; Roger, S.F.; Luo, X.Z. Allelopathically inhibitory effects of eucalyptus extracts on the growth of Microcystis aeruginosa. Chemosphere 2019, 225, 424–433. [Google Scholar] [CrossRef]
- Yu, Y.; Zeng, Y.D.; Li, J.; Yang, C.Y.; Zhang, X.H.; Luo, F.; Dai, X.Z. An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total Environ. 2019, 650, 34–43. [Google Scholar] [CrossRef]
- Arora, A.; Sairam, R.K.; Srivastava, G.C. Oxidative stress and antioxidative system in plants. Curr. Sci. 2002, 82, 1227–1239. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayali, X.; Azam, F. Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 2004, 51, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.N.; Ding, N.; Gao, P.K.; Han, M.X.; Liu, X.X.; Wang, J.G.; Li, S.; Fu, B.Y.; Wang, R.J.; Zhou, J. Diverse algicidal bacteria associated with harmful bloom-forming Karenia mikimotoi in estuarine soil and seawater. Sci. Total Environ. 2018, 631, 1415–1420. [Google Scholar] [CrossRef]
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.H.; Zhou, B.; Xu, L.; Liu, L.L.; Wang, G.Y.; Liu, X.D.; Tang, X.X. A marine algicidal Thalassospira and its active substance against the harmful algal bloom species Karenia mikimotoi. Appl. Microbiol. Biotechnol. 2016, 100, 5131–5139. [Google Scholar] [CrossRef]
- Hou, S.L.; Shu, W.J.; Tan, S.; Zhao, L.; Yin, P.H. Exploration of the antioxidant system and photosynthetic system of a marine algicidal Bacillus and its effect on four harmful algal bloom species. Can. J. Microbiol. 2016, 62, 49–59. [Google Scholar] [CrossRef]
- Hu, X.L.; Yin, P.H.; Zhao, L.; Yu, Q.M. Characterization of cell viability in Phaeocystis globosa cultures exposed to marine algicidal bacteria. Biotechnol. Bioprocess Eng. 2015, 20, 58–66. [Google Scholar] [CrossRef]
- Shao, J.H.; He, Y.X.; Chen, A.W.; Peng, L.; Luo, S.; Wu, G.Y.; Zou, H.L.; Li, R.H. Interactive effects of algicidal efficiency of Bacillus sp. B50 and bacterial community on susceptibility of Microcystis aeruginosa with different growth rates. Int. Biodeterior. Biodegrad. 2015, 97, 1–6. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Ishida, K.; Ito, Y.; Okada, S.; Murakami, M. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Lett. 2003, 44, 8005–8007. [Google Scholar] [CrossRef]
- Wu, L.M.; Wu, H.J.; Chen, L.N.; Xie, S.S.; Zang, H.Y.; Borriss, R.; Gao, X.W. Bacilysin from Bacillus amyloliquefaciens FZB42 has specific bactericidal activity against harmful algal bloom species. Appl. Environ. Microbiol. 2014, 80, 7512–7520. [Google Scholar] [CrossRef] [Green Version]
- Skerratt, J.H.; Bowman, J.P.; Hallegraeff, G.; James, S.; Nichols, P.D. Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar. Ecol. Prog. Ser. 2002, 244, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.Y.; Keesing, J.K.; Xing, Q.G.; Shi, P. World’s largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Mar. Pollut. Bull. 2009, 58, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Xiao, J.; Fan, S.L.; Li, Y.; Liu, X.Q.; Liu, D.Y. Who made the world’s largest green tide in China?—An integrated study on the initiation and early development of the green tide in Yellow Sea. Limnol. Oceanogr. 2015, 60, 1105–1117. [Google Scholar] [CrossRef]
- Ye, N.H.; Zhuang, Z.Z.; Jin, X.; Wang, Q.; Zhang, X.; Li, D.M.; Wang, H.X.; Mao, Y.Z.; Jiang, Z.J.; Li, B.; et al. China is on the track tackling Enteromorpha spp forming green tide. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Ye, N.H.; Zhang, X.W.; Mao, Y.Z.; Liang, C.W.; Xu, D.; Zou, J.; Zhuang, Z.Z.; Wang, Q.Y. ‘Green tides’ are overwhelming the coastline of our blue planet: Taking the world’s largest example. Ecol. Res. 2011, 26, 477–485. [Google Scholar] [CrossRef]
- Huo, Y.Z.; Han, H.B.; Shi, H.H.; Wu, H.L.; Zhang, J.H.; Yu, K.F.; Xu, R.; Liu, C.C.; Zhang, Z.L.; Liu, K.F.; et al. Changes to the biomass and species composition of Ulva sp. on Porphyra aquaculture rafts, along the coastal radial sandbank of the Southern Yellow Sea. Mar. Pollut. Bull. 2015, 93, 210–216. [Google Scholar] [CrossRef]
- Zhang, J.H.; Huo, Y.Z.; Wu, H.; Yu, K.; Kim, J.K.; Yarish, C.; Qin, Y.T.; Liu, C.C.; Xu, R.; He, P.M. The origin of the Ulva macroalgal blooms in the Yellow Sea in 2013. Mar. Pollut. Bull. 2014, 89, 276–283. [Google Scholar] [CrossRef]
- Li, H.M.; Zhang, Y.Y.; Chen, J.; Zheng, X.; Liu, F.; Jiao, N.Z. Nitrogen uptake and assimilation preferences of the main green tide alga Ulva prolifera in the Yellow Sea, China. J. Appl. Phycol. 2018, 31, 625–635. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, X.H.; Gao, C.L.; Jiang, M.J.; Li, R.X.; Wang, Z.L.; Li, Y.; Fan, S.L.; Zhang, X.L. Effect of temperature, salinity and irradiance on growth and photosynthesis of Ulva prolifera. Acta Oceanol. Sin. 2016, 35, 114–121. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, T.; Yu, R.C.; Zhang, Q.C.; Zhou, M.J. Interactions between selected microalgae and microscopic propagules of Ulva prolifera. J. Mar. Biol. Assoc. UK 2017, 98, 1571–1580. [Google Scholar] [CrossRef]
- Fan, X.; Xu, D.; Wang, Y.T.; Zhang, X.W.; Cao, S.N.; Mou, S.L.; Ye, N.H. The effect of nutrient concentrations, nutrient ratios and temperature on photosynthesis and nutrient uptake by Ulva prolifera: Implications for the explosion in green tides. J. Appl. Phycol. 2014, 26, 537–544. [Google Scholar] [CrossRef]
- Sun, X.; Wu, M.Q.; Xing, Q.G.; Song, X.D.; Zhao, D.H.; Han, Q.Q.; Zhang, G.Z. Spatio-temporal patterns of Ulva prolifera blooms and the corresponding influence on chlorophyll-a concentration in the Southern Yellow Sea, China. Sci. Total Environ. 2018, 640, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Springer: Boston, MA, USA, 1975. [Google Scholar]
- Jin, Q.; Dong, S.L.; Wang, C.Y. Allelopathic growth inhibition of Prorocentrum micans (Dinophyta) by Ulva pertusa and Ulva linza (Chlorophyta) in laboratory cultures. Eur. J. Phycol. 2005, 40, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Huang, H.J.; Li, H.Y.; Liu, J.S.; Yang, W.D. Genetic diversity of Ulva prolifera population in Qingdao coastal water during the green algal blooms revealed by: Microsatellite. Mar. Pollut. Bull. 2016, 111, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sun, X.; Lu, Z.; Liu, B.; Zhou, Q.; Zhang, Y.; Wu, Z. Allelopathic effects of pyrogallic acid secreted by submerged macrophytes on Microcystis aeruginosa: Role of ROS generation. Allelopath. J. 2014, 33, 121–130. [Google Scholar]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Tang, X.X.; Zhang, H.; Qu, T.F.; Wang, Y. Photosynthetic adaptation strategy of Ulva prolifera floating on the sea surface to environmental changes. Plant Physiol. Biochem. 2016, 107, 116–125. [Google Scholar] [CrossRef]
- Wang, J.W.; Yan, B.L.; Lin, A.P.; Hu, J.P.; Shen, S.D. Ecological factor research on the growth and induction of spores release in Enteromorpha Prolifera (Chlorophyta). Mar. Sci. Bull. 2007, 26, 60–66. [Google Scholar]
- Zhao, P.J.; Wang, H.X.; Li, G.H.; Li, H.D.; Liu, J.; Shen, Y.M. Secondary metabolites from endophytic Streptomyces sp. Lz531. Chem. Biodivers. 2007, 4, 899–904. [Google Scholar] [CrossRef]
- Li, T.; Wang, G.C.; Huang, X.J.; Ye, W.C. ChemInform Abstract: Whitmanoside A (I), a New α-Pyrone Glycoside from the Leech Whitmania pigra. J. Cheminform. 2013, 44. [Google Scholar] [CrossRef]
- Furtado, N.A.J.C.; Pupo, M.T.; Carvalho, I.; Campo, V.L.; Duarte, M.C.T.; Bastos, J.K. Diketopiperazines produced by an Aspergillus fumigatus Brazilian strain. J. Braz. Chem. Soc. 2005, 16, 1448–1453. [Google Scholar] [CrossRef]
- Tilney, C.L.; Pokrzywinski, K.L.; Coyne, K.J.; Warner, M.E. Effects of a bacterial algicide, IRI-160AA, on dinoflagellates and the microbial community in microcosm experiments. Harmful Algae 2014, 39, 210–222. [Google Scholar] [CrossRef]
- Meyer, N.; Bigalke, A.; Kaulfuss, A.; Pohnert, G. Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 2017, 41, 880–899. [Google Scholar] [CrossRef] [Green Version]
- Hare, C.E.; Demir, E.; Coyne, K.J.; Craig Cary, S.; Kirchman, D.L.; Hutchins, D.A. A bacterium that inhibits the growth of Pfiesteria piscicida and other dinoflagellates. Harmful Algae 2005, 4, 221–234. [Google Scholar] [CrossRef]
- Zozaya-Valdes, E.; Egan, S.; Thomas, T. A comprehensive analysis of the microbial communities of healthy and diseased marine macroalgae and the detection of known and potential bacterial pathogens. Front. Microbiol. 2015, 6, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Perveen, S.; Mushtaq, M.N.; Yousaf, M.; Sarwar, N. Allelopathic hormesis and potent allelochemicals from multipurpose tree Moringa oleifera leaf extract. Plant Biosyst. 2020, 18, 1–6. [Google Scholar] [CrossRef]
- Wang, C.X.; Zhu, M.X.; Chen, X.H.; Qu, B. Review on allelopathy of exotic invasive plants. Procedia Eng. 2011, 18, 240–246. [Google Scholar]
- Li, N.C.; Tong, M.M.; Glibert, P.M. Effect of allelochemicals on photosynthetic and antioxidant defense system of Ulva prolifera. Aquat. Toxicol. 2020, 224, 105513. [Google Scholar] [CrossRef]
- Xu, D.; Gao, Z.Q.; Zhang, X.W.; Fan, X.; Wang, Y.T.; Li, D.M.; Wang, W.; Zhuang, Z.; Ye, N. Allelopathic interactions between the opportunistic species Ulva prolifera and the native macroalga Gracilaria lichvoides. PLoS ONE 2012, 7, e33648. [Google Scholar] [CrossRef]
- Zhou, Q.X.; Hu, X.G. Systemic stress and recovery patterns of rice roots in response to graphene oxide nanosheets. Environ. Sci. Technol. 2017, 51, 2022–2030. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, X.Y.; Tang, X.X. Antioxidant system responses in two co-occurring green-tide algae under stress conditions. J. Ocean Univ. 2016, 34, 102–108. [Google Scholar] [CrossRef]
- Tan, S.; Hu, X.L.; Yin, P.H.; Zhao, L. Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism. J. Microbiol. 2016, 54, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Liu, X.L.; Pan, J.L.; Yang, H. Synergistic algicidal effect and mechanism of two diketopiperazines produced by Chryseobacterium sp. strain GLY-1106 on the harmful bloom-forming Microcystis aeruginosa. Sci. Rep. 2015, 5, 14720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.X.; Xu, J.R.; Cheng, Y. Quantitative analyses of relationships between ecotoxicological effects and combined pollution. Plant Soil 2004, 261, 155–162. [Google Scholar] [CrossRef]
- Hess, F.D. Light-dependent herbicides: An overview. Weed Sci. 2000, 48, 160–170. [Google Scholar] [CrossRef]
- Ni, L.T.; Rong, S.Y.; Gu, G.X.; Hu, L.L.; Wang, P.F.; Li, D.Y.; Yue, F.F.; Wang, N.; Wu, H.Q.; Li, S.Y. Inhibitory effect and mechanism of linoleic acid sustained-release microspheres on Microcystis aeruginosa at different growth phases. Chemosphere 2018, 212, 654–661. [Google Scholar] [CrossRef]
- Wang, G.X.; Zhang, Q.; Li, J.L.; Chen, X.Y.; Lang, Q.L.; Kuang, S.P. Combined effects of erythromycin and enrofloxacin on antioxidant enzymes and photosynthesis-related gene transcription in Chlorella vulgaris. Aquat. Toxicol. 2019, 212, 138–145. [Google Scholar] [CrossRef]
- Zhou, Q.X.; Yue, Z.K.; Li, Q.Z.; Zhou, R.R.; Liu, L. Exposure to PbSe nanoparticles and male reproductive damage in a rat model. Environ. Sci. Technol. 2019, 53, 13408–13416. [Google Scholar] [CrossRef] [Green Version]
- Kurama, E.E.; Fenille, R.C.; Rosa, V.E., Jr.; Rosa, D.D.; Ulian, E.C. Mining the enzymes involved in the detoxification of reactive oxygen species (ROS) in sugarcane. Mol. Plant Pathol. 2010, 3, 251–259. [Google Scholar] [CrossRef]
- Fan, M.H.; Sun, X.; Xu, N.J.; Liao, Z.; Wang, R.X. cDNA cloning, characterization and expression analysis of manganese superoxide dismutase in Ulva prolifera. J. Appl. Phycol. 2015, 28, 1391–1401. [Google Scholar] [CrossRef]
- Cruces, E.; Rautenberger, R.; Cubillos, V.M.; Ramirez-Kushel, E.; Rojas-Lillo, Y.; Lara, C.; Montory, J.A.; Gomez, I. Interaction of photoprotective and acclimation mechanisms in Ulva rigida (Chlorophyta) in response to diurnal changes in solar radiation in Southern Chile. J. Phycol. 2019, 55, 1011–1027. [Google Scholar] [CrossRef]
- Sung, M.S.; Hsu, Y.T.; Wu, T.M.; Lee, T.M. Hypersalinity and hydrogen peroxide upregulation of gene expression of antioxidant enzymes in Ulva fasciata against oxidative stress. Mar. Biotechnol. 2009, 11, 199–209. [Google Scholar] [CrossRef]
- Xu, S.; Yang, S.Q.; Yang, Y.J.; Xu, J.Z.; Shi, J.Q.; Wu, Z.X. Influence of linoleic acid on growth, oxidative stress and photosynthesis of the cyanobacterium Cylindrospermopsis raciborskii. N. Z. J. Mar. Freshw. Res. 2017, 51, 223–236. [Google Scholar] [CrossRef]
- Fan, M.H.; Sun, X.; Liao, Z.; Wang, J.X.; Cui, D.L.; Xu, N.J. Full-length cDNA cloning, characterization of catalase from Ulva prolifera and antioxidant response to diphenyliodonium. J. Appl. Phycol. 2018, 30, 3361–3372. [Google Scholar] [CrossRef]
- Long, M.; Tallec, K.; Soudant, P.; Le Grand, F.; Donval, A.; Lambert, C.; Sarthou, G.; Jolley, D.F.; Hégaret, H. Allelochemicals from Alexandrium minutum induce rapid inhibition of metabolism and modify the membranes from Chaetoceros muelleri. Algal Res. 2018, 35, 508–518. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Szeto, Y.T.; Jiang, C.; Wang, X.; Tao, Y.; Tu, J.; Chen, J. Effects of Dracontomelon duperreanum leaf litter on the growth and photosynthesis of Microcystis aeruginosa. Bull. Environ. Contam. Toxicol. 2018, 100, 690–694. [Google Scholar] [CrossRef]
- Yu, S.M.; Li, C.; Xu, C.C.; Effiong, K.; Xiao, X. Understanding the inhibitory mechanism of antialgal allelochemical flavonoids from genetic variations: Photosynthesis, toxin synthesis and nutrient utility. Ecotox. Environ. Saf. 2019, 177, 18–24. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Zheng, Z.Z.; Gao, S.; Wang, G.C. Far red light induces the expression of LHCSR to trigger nonphotochemical quenching in the intertidal green macroalgae Ulva prolifera. Algal Res. 2019, 40, 101512. [Google Scholar] [CrossRef]
- Gao, S.; Shen, S.D.; Wang, G.C.; Niu, J.F.; Lin, A.P.; Pan, G.H. PSI-driven cyclic electron flow allows intertidal macro-algae Ulva sp. (Chlorophyta) to survive in desiccated conditions. Plant Cell Physiol. 2011, 52, 885–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Chi, Z.; Chen, H.L.; Zheng, Z.B.; Weng, Y.X.; Wang, G.C. A Supercomplex, of approximately 720 kDa and composed of both photosystem reaction centers, dissipates excess energy by PSI in green macroalgae under salt stress. Plant Cell Physiol. 2019, 60, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Lelong, A.; Haberkorn, H.; Le Goïc, N.; Hégaret, H.; Soudant, P. A new insight into allelopathic effects of Alexandrium minutum on photosynthesis and respiration of the diatom Chaetoceros neogracile revealed by photosynthetic-performance analysis and flow cytometry. Microb. Ecol. 2011, 62, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Genty, B.; Briantais, J.M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Mhatre, A.; Patil, S.; Agarwal, A.; Pandit, R.; Lali, A.M. Influence of nitrogen source on photochemistry and antenna size of the photosystems in marine green macroalgae, Ulva lactuca. Photosynth. Res. 2019, 139, 539–551. [Google Scholar] [CrossRef]
- Peers, G.; Truong, T.B.; Ostendorf, E.; Busch, A.; Elrad, D.; Grossman, A.R.; Hippler, M.; Niyogi, K.K. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 2009, 462, 518–521. [Google Scholar] [CrossRef]
- Figueroa, F.L.; Celis-Plá, P.S.M.; Martínez, B.; Korbee, N.; Trilla, A.; Arenas, F. Yield losses and electron transport rate as indicators of thermal stress in Fucus serratus (Ochrophyta). Algal Res. 2019, 41, 101560. [Google Scholar] [CrossRef]
- Dong, M.T.; Zhang, X.W.; Zhuang, Z.Z.; Zou, J.; Ye, N.H.; Xu, D.; Mou, S.L.; Liang, C.W.; Wang, W.Q. Characterization of the LhcSR gene under light and temperature stress in the green alga Ulva linza. Plant Mol. Biol. Rep. 2011, 30, 10–16. [Google Scholar] [CrossRef]
- Mou, S.L.; Zhang, X.W.; Dong, M.; Fan, X.; Xu, J.; Cao, S.; Xu, D.; Wang, W.; Ye, N.H. Photoprotection in the green tidal alga Ulva prolifera: Role of LhcSR and PsbS proteins in response to high light stress. Plant Biol. 2013, 15, 1033–1039. [Google Scholar] [CrossRef]
- Kommalapati, M.; Hwang, H.J.; Wang, H.L.; Burnap, R.L. Engineered ectopic expression of the psbA gene encoding the photosystem II D1 protein in Synechocystis sp. PCC6803. Photosynth. Res. 2007, 92, 315–325. [Google Scholar] [CrossRef]
- Barati, B.; Lim, P.E.; Gan, S.Y.; Poong, S.W.; Phang, S.M. Gene expression profile of marine Chlorella strains from different latitudes: Stress and recovery under elevated temperatures. J. Appl. Phycol. 2018, 30, 3121–3130. [Google Scholar] [CrossRef]
- Ohnishi, N.; Allakhverdiev, S.I.; Takahashi, S.; Higashi, S.; Watanabe, M.; Nishiyama, Y.; Norio, M. Two-step mechanism of photodamage to photosystem II: Step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 2005, 44, 8494–8499. [Google Scholar] [CrossRef] [PubMed]
- Hakala, M.; Tuominen, I.; Keränen, M.; Tyystjärvi, T.; Tyystjärvi, E. Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of Photosystem II. Biochim. Biophys. Acta Bioenergy 2005, 1706, 68–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa-Galvis, V.; Redekop, P.; Guan, K.; Griess, A.; Truong, T.B.; Wakao, S.; Niyogi, K.K.; Jahns, P. Photosystem II Subunit PsbS is involved in the induction of LHCSR protein-dependent energy dissipation in Chlamydomonas reinhardtii. J. Biol. Chem. 2016, 291, 17478–17487. [Google Scholar] [CrossRef] [Green Version]
- Pinnola, A.; Cazzaniga, S.; Alboresi, A.; Nevo, R.; Levin-Zaidman, S.; Reich, Z.; Bassi, R. Light-Harvesting Complex stress-eelated proteins catalyze excess energy dissipation in both photosystems of physcomitrella patens. Plant Cell 2015, 27, 3213–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhu, H.; Lei, X.; Zhang, H.; Cai, G.; Chen, Z.; Fu, L.; Xu, H.; Zheng, T.L. The death mechanism of the harmful algal bloom species Alexandrium tamarense induced by algicidal bacterium deinococcus sp. Y35. Front. Microbiol. 2015, 6, 992–997. [Google Scholar] [CrossRef] [Green Version]
- Perzborn, M.; Syldatk, C.; Rudat, J. Enzymatical and microbial degradation of cyclic dipeptides (diketopiperazines). AMB Express 2013, 3, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Primer | Sequence (5′–3′) |
|---|---|
| 5S | F: 5′-GGTTGGGCAGGATTAGTA-3′ |
| R: 5′-AGGCTTAAGTTGCGAGTT-3′ | |
| ITS | F: 5′-TCGTAACAAGGTTTCCGTAGG-3′ |
| R: 5′-GCTGCGTTCTTCATCGWTG-3′ |
| Primer | Sequence (5′-3′) | Product Length |
|---|---|---|
| Tubulin | F: 5′-CAAGGATGTCAATGCTGCTGT-3′ | 112 |
| R: 5′-GACCGTAGGTGGCTGGTAGTT-3′ | ||
| PsbS | F: 5′-AACAGGTTCATCCATCACGG-3′ | 121 |
| R: 5′-TTGCCTCAAACTCATCCTCTG-3′ | ||
| LhcSR | F: 5′-CTATGCGAAGACTCTCAACG-3′ | 83 |
| R: 5′-CCTCGCGGTAGCGCTTAACT-3′ | ||
| PsbA | F: 5′- CTTTATGGGCTCGCTTTTGT-3′ | 103 |
| R: 5′- TGGAACTACAGCACCAGAAA-3′ | ||
| PsbD | F: 5′- CAGGAAGTGTTCAACCAGTA-3′ | 167 |
| R: 5′- AGCAGCGATGTGATGAGACG-3′ | ||
| upMnSOD | F: 5′-ATCACCAGGCGTATGTCACC-3′ | 94 |
| R: 5′-TTCAAGTGCCCTCCACCGTT-3′ | ||
| upCAT | F: 5′-CTCTCAAGCCCAATCCTCGT-3′ | 95 |
| R: 5′-AGTTCAGTGGGATGCCAACA-3′ |
| Parameter | Definition | Equation |
|---|---|---|
| Fv/Fm | maximum quantum yield of PSII | (Fm − F0)/Fm |
| Y(II) | effective quantum yield of PSII | (F’m − Ft)/F’m |
| NPQ | non-photochemical quenching | (Fm − F’m)/F’m |
| rETR | relative electron transport rate | 0.5 × Y(II) × PAR × IA |
| qP | photochemical quenching | (F’m − Ft)/(F’m − F’0) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Zhang, J.; Zhao, X.; Wang, P.; Tong, M.; Glibert, P.M. Allelopathic Inhibition by the Bacteria Bacillus cereus BE23 on Growth and Photosynthesis of the Macroalga Ulva prolifera. J. Mar. Sci. Eng. 2020, 8, 718. https://doi.org/10.3390/jmse8090718
Li N, Zhang J, Zhao X, Wang P, Tong M, Glibert PM. Allelopathic Inhibition by the Bacteria Bacillus cereus BE23 on Growth and Photosynthesis of the Macroalga Ulva prolifera. Journal of Marine Science and Engineering. 2020; 8(9):718. https://doi.org/10.3390/jmse8090718
Chicago/Turabian StyleLi, Naicheng, Jingyao Zhang, Xinyu Zhao, Pengbin Wang, Mengmeng Tong, and Patricia M. Glibert. 2020. "Allelopathic Inhibition by the Bacteria Bacillus cereus BE23 on Growth and Photosynthesis of the Macroalga Ulva prolifera" Journal of Marine Science and Engineering 8, no. 9: 718. https://doi.org/10.3390/jmse8090718