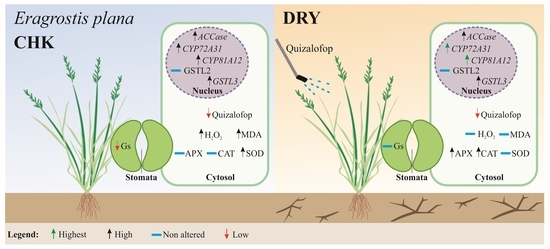
Transgenerational Effect of Drought Stress and Sub-Lethal Doses of Quizalofop-p-ethyl: Decreasing Sensitivity to Herbicide and Biochemical Adjustment in Eragrostis plana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Establishment of the Populations
2.2. Dose-Response Curves with Quizalofop
2.3. Physiological and Biochemical Analyses and Quizalofop-p Concentration of G2 Populations
Variables Measured
2.4. Concentration of Quizalofop and Molecular Analyses in G2 Populations
2.4.1. Quizalofop Quantitation
2.4.2. Gene Expression
2.5. Statistics Analyses
3. Results and Discussion
3.1. Dose-Response Curves Assessing Sensitivity to Quizalofop
3.2. Stomatal Conductance
3.3. H2O2 Content and Lipid Peroxidation
3.4. Antioxidant Enzymes
3.5. Quizalofop Concentration
3.6. Gene Expression
3.7. Mechanisms Involving Reducing Sensitivity Responses to Quizalofop in E. plana
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guido, A.; Hoss, D.; Pillar, V.D. Competitive effects and responses of the invasive grass Eragrostis plana in Río de la Plata grasslands. Austral. Ecol. 2019, 44, 1478–1486. [Google Scholar] [CrossRef]
- Medeiros, R.B.; Focht, T. Invasão, prevenção, controle e utilização do capim-annoni-2 (Eragrostis plana Nees) no Rio Grande do Sul, Brasil. Pesqui. Agropecuária Gaúcha 2007, 13, 105–114. [Google Scholar]
- Barbosa, F.G.; Pillar, V.D.; Palmer, A.R.; Melo, A.S. Predicting the current distribution and potential spread of the exotic grass Eragrostis plana Nees in South America and identifying a bioclimatic niche shift during invasion. Austral. Ecol. 2013, 38, 260–267. [Google Scholar] [CrossRef]
- Bastiani, M.O.; Roma-Burgos, N.; Langaro, A.C.; Salas-Perez, R.A.; Rouse, C.E.; Fipke, M.V.; Lamego, F.P. Ammonium sulfate improves the efficacy of glyphosate on south african lovegrass (Eragrostis plana) under water stress. Weed Sci. 2021, 2507, 1–43. [Google Scholar] [CrossRef]
- Carloto, B.W.; Buriol, G.A.; Dornelles, S.H.B.; Trivisiol, V.S.; Peripolli, M.; Escobar, O.S. Morphological and phenological responses of Eragrostis plana Nees and Eragrostis pilosa (L.) P. Beauv. plants subjected to different soil moisture conditions. Planta Daninha 2019, 37, 1–7. [Google Scholar] [CrossRef]
- Favaretto, A.; Santos, J.; Carneiro, C.M.; Basso, S.M.S. The first anatomical and histochemical study of tough lovegrass (Eragrostis plana Nees, Poaceae). Afr. J. Agric. Res. 2015, 10, 2940–2947. [Google Scholar] [CrossRef]
- Dyer, W.E. Stress-induced evolution of herbicide resistance and related pleiotropic effects. Pest. Manag. Sci. 2018, 74, 1759–1768. [Google Scholar] [CrossRef]
- Kleinman, Z.; Ben-Ami, G.; Rubin, B. From sensitivity to resistance—Factors affecting the response of Conyza spp. to glyphosate. Pest. Manag. Sci. 2016, 72, 1681–1688. [Google Scholar] [CrossRef]
- Huan, Z.B.; Zhang, H.J.; Zhen, H.O.U.; Zhang, S.Y.; Zhang, Y.; Liu, W.; Bi, Y.; Wang, J. Resistance level and metabolism of barnyard-grass (Echinochloa crusgalli (L.) Beauv.) populations to quizalofop-p-ethyl in heilongjiang province, China. Agric. Sci. China 2011, 10, 1914–1922. [Google Scholar] [CrossRef]
- Luo, X.; Liu, Z.; Sunohara, Y.; Matsumoto, H.; Li, P. Involvement of H2O2 in fluazifop-p-butyl-induced cell death in bristly starbur seedlings. Pestic. Biochem. Physiol. 2017, 143, 258–264. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant. Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, Z.; Xu, Z.; Lv, D.; Wang, J. Determination of ACCase sensitivity and gene expression in quizalofop–ethyl resistant and susceptible barnyardgrass (Echinochloa crus-galli) biotypes. Weed Sci. 2013, 61, 537–542. [Google Scholar] [CrossRef]
- Dayan, F.E.; Barker, A.; Bough, R.; Ortiz, M.; Takano, H.; Duke, S.O. Herbicide mechanisms of action and resistance. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 4, pp. 36–48. [Google Scholar]
- Takano, H.K.; Ovejero, R.F.L.; Belchior, G.G.; Maymone, G.P.L.; Dayan, F.E. ACCase-inhibiting herbicides: Mechanism of action, resistance evolution and stewardship. Sci. Agric. 2020, 78, 1–11. [Google Scholar] [CrossRef]
- Radwan, D.E.M. Salicylic acid induced alleviation of oxidative stress caused by clethodim in maize (Zea mays L.) leaves. Pestic. Biochem. Physiol. 2012, 102, 182–188. [Google Scholar] [CrossRef]
- Benedetti, L.; Rangani, G.; Ebeling Viana, V.; Carvalho-Moore, P.; Rabaioli Camargo, E.; de Avila, L.A.; Roma-Burgos, N. Recurrent selection by herbicide sublethal dose and drought stress results in rapid reduction of herbicide sensitivity in junglerice. Agronomy 2020, 10, 1619. [Google Scholar] [CrossRef]
- Galloway, L.F. Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytol. 2005, 166, 93–100. [Google Scholar] [CrossRef]
- Whittle, C.A.; Otto, S.P.; Johnston, M.O.; Krochko, J.E. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 2009, 87, 650–657. [Google Scholar] [CrossRef] [Green Version]
- Nosalewicz, A.; Siecińska, J.; Śmiech, M.; Nosalewicz, M.; Wiącek, D.; Pecio, A.; Wach, D. Transgenerational effects of temporal drought stress on spring barley morphology and functioning. Environ. Exp. Bot. 2016, 131, 120–127. [Google Scholar] [CrossRef]
- Busi, R.; Neve, P.; Powles, S. Evolved polygenic herbicide resistance in Lolium rigidum by low-dose herbicide selection within standing genetic variation. Evol. Appl. 2013, 6, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest. Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef] [PubMed]
- Rigon, C.A.G.; Gaines, T.A.; Küpper, A.; Dayan, F.E. Metabolism-based herbicide resistance, the major threat among the non-target site resistance mechanisms. Outlooks Pest. Manag. 2020, 31, 162–168. [Google Scholar] [CrossRef]
- Li, W.; Zhang, L.; Zhao, N.; Guo, W.; Liu, W.; Li, L.; Wang, J. Multiple resistance to accase and ALS-inhibiting herbicides in Beckmannia syzigachne (Steud.) fernald without mutations in the target enzymes. Chil. J. Agric. Res. 2017, 77, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants protective role of exogenous polyamines. Plant. Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 2, 248–254. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. occurrence in higher plants. Plant. Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Havir, E.A.; McHale, N.A. Biochemical and developmental characterization of multiple forms of catalase in Tobacco leaves. Plant. Physiol. 1987, 84, 450–455. [Google Scholar] [CrossRef] [Green Version]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant. Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Li, L.; Li, W.; Qin, D.; Jiang, S.; Liu, F. Application of graphitized carbon black to the QuEChERS method for pesticide Multiresidue analysis in spinach. J. AOAC Int. 2009, 92, 538–547. [Google Scholar] [CrossRef] [Green Version]
- Laforest, M.; Soufiane, B.; Simard, M.J.; Obeid, K.; Page, E.; Nurse, R.E. Acetyl-CoA carboxylase overexpression in herbicide-resistant large crabgrass (Digitaria sanguinalis). Pest. Manag. Sci. 2017, 73, 2227–2235. [Google Scholar] [CrossRef]
- Chen, G.; Xu, H.; Zhang, T.; Bai, C.; Dong, L. Fenoxaprop-P-ethyl resistance conferred by cytochrome P450s and target site mutation in Alopecurus japonicus. Pest. Manag. Sci. 2018, 74, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 2019, 221, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant. Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saika, H.; Horita, J.; Taguchi-Shiobara, F.; Nonaka, S.; Nishizawa-Yokoi, A.; Iwakami, S.; Hori, K.; Matsumoto, T.; Tanaka, T.; Itoh, T.; et al. A novel rice cytochrome P450 gene, CYP72A31, confers tolerance to acetolactate synthase-inhibiting herbicides in rice and arabidopsis. Plant. Physiol. 2014, 166, 1232–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummins, I.; Wortley, D.J.; Sabbadin, F.; He, Z.; Coxon, C.R.; Straker, H.E.; Sellars, J.D.; Knight, K.; Edwards, L.; Hughes, D.; et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA 2013, 110, 5812–5817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Lu, D.; Li, C.; Luo, J.; Zhu, B.F.; Zhu, J.; Shangguan, Y.; Wang, Z.; Sang, T.; Zhou, B.; et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor Shattering Abortion1. Plant. Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- NCBI National Center for Biotechnology Information. Available online: www.ncbi.nlm.nih.gov (accessed on 10 January 2020).
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant. Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Rocha Pereira, M.R.; Sasso, F.; de Souza, G.; Iran Cardoso da Silva, J.; Macedo, A.C.; Martins, D. Influence of soil water potential in the action of herbicides on goosegrass (Eleusine indica (L) Gaertn). Biosci. J. 2015, 31, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Manalil, S.; Busi, R.; Renton, M.; Powles, S.B. Rapid evolution of herbicide resistance by low herbicide dosages. Weed Sci. 2011, 59, 210–217. [Google Scholar] [CrossRef]
- Neve, P.; Powles, S. Recurrent selection with reduced herbicide rates results in the rapid evolution of herbicide resistance in Lolium rigidum. Theor. Appl. Genet. 2005, 110, 1154–1166. [Google Scholar] [CrossRef]
- Ribeiro, R.V.; Machado, R.S.; Machado, E.C.; Machado, D.F.S.P.; Magalhães Filho, J.R.; Landell, M.G.A. Revealing drought-resistance and productive patterns in sugarcane genotypes by evaluating both physiological responses and stalk yield. Exp. Agric. 2013, 49, 212–224. [Google Scholar] [CrossRef]
- Nascentes, R.F.; Carbonari, C.A.; Simões, P.S.; Brunelli, M.C.; Velini, E.D.; Duke, S.O. Low doses of glyphosate enhance growth, CO2 assimilation, stomatal conductance and transpiration in sugarcane and eucalyptus. Pest. Manag. Sci. 2018, 74, 1197–1205. [Google Scholar] [CrossRef]
- Jang, S.; Marjanovic, J.; Gornicki, P. Resistance to herbicides caused by single amino acid mutations in acetyl-CoA carboxylase in resistant populations of grassy weeds. New Phytol. 2013, 197, 1110–1116. [Google Scholar] [CrossRef]
- Banas, A.; Johasson, I.; Stenlid, G.; Stymne, S. Free-radical scavengers and inhibitors of lipoxygenases as antagonists against the herbicides haloxyfop and alloxydim. Swedish J. Agric. Res. 1993, 23, 67–75. [Google Scholar]
- Shang, Y.; Yang, C.; Liu, Z.; Song, J.; Li, P.; Li, L.; Zhou, F.; Xin, H.; Wan, F.; Matsumoto, H.; et al. New evidence for primordial action site of fluazifop-p-butyl on Acanthospermum hispidum seedlings: From the effects on chlorophyll fluorescence characteristics and histological observation. Pestic. Biochem. Physiol. 2017, 142, 170–175. [Google Scholar] [CrossRef]
- Soares, A.R.; de Ferrarese, M.L.L.; de Siqueira-Soares, R.C.; Marchiosi, R.; Finger-Teixeira, A.; Ferrarese-Filho, O. The allelochemical L-DOPA increases melanin production and reduces reactive oxygen species in soybean roots. J. Chem. Ecol. 2011, 37, 891–898. [Google Scholar] [CrossRef]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, K.W.; Wu, J.; Hatzios, K.K.; Hagood, E.S. The mechanism of resistance to aryloxyphenoxypropionate and cyclohexanedione herbicides in a johnsongrass biotype. Weed Sci. 2001, 49, 477–484. [Google Scholar] [CrossRef]
- Tardif, F.J.; Leroux, G.D. Translocation of glyphosate and quizalofop and metabolism of quizalofop in quackgrass biotypes (Elytrigia repens). Weed Technol. 1991, 5, 525–531. [Google Scholar] [CrossRef]
- Koeppe, M.K.; Anderson, J.J.; Shalaby, L.M. Metabolism of [14C]quizalofop-ethyl in soybean and cotton plants. J. Agric. Food Chem. 1990, 38, 1085–1091. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, Y.; Xia, T.; Chen, J.; Cai, X. Differential enantioselectivity of quizalofop ethyl and its acidic metabolite: Direct enantiomeric separation and assessment of multiple toxicological endpoints. J. Hazard. Mater. 2011, 186, 876–882. [Google Scholar] [CrossRef]
- López-Ruiz, R.; Romero-González, R.; Martínez Vidal, J.L.; Fernández-Pérez, M.; Garrido Frenich, A. Degradation studies of quizalofop-p and related compounds in soils using liquid chromatography coupled to low and high resolution mass analyzers. Sci. Total Environ. 2017, 607–608, 204–213. [Google Scholar] [CrossRef]
- Iwakami, S.; Uchino, A.; Kataoka, Y.; Shibaike, H.; Watanabe, H.; Inamura, T. Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest. Manag. Sci. 2014, 70, 549–558. [Google Scholar] [CrossRef]
- Gaines, T.A.; Lorentz, L.; Figge, A.; Herrmann, J.; Maiwald, F.; Ott, M.C.; Han, H.; Busi, R.; Yu, Q.; Powles, S.B.; et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant. J. 2014, 78, 865–876. [Google Scholar] [CrossRef]
- Chen, W.; Wu, L.; Wang, J.; Yu, Q.; Bai, L.; Pan, L. Quizalofop-p-ethyl resistance in Polypogon fugax involves glutathione. Pest. Manag. Sci. 2020, 76, 3800–3805. [Google Scholar] [CrossRef]
- Pégeot, H.; Koh, C.S.; Petre, B.; Mathiot, S.; Duplessis, S.; Hecker, A.; Didierjean, C.; Rouhier, N. The poplar Phi class glutathione transferase: Expression, activity and structure of GSTF1. Front. Plant. Sci. 2014, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]







| Target Gene | Forward (5′-3′) | Reverse (5′-3′) | Reference |
|---|---|---|---|
| OsACT1 | CCTTCAACACCCCTGCTATG | CAATGCCAGGGAACATAGTG | [37] |
| Os18S | CTACGTCCCTGCCCTTTGTACA | ACACTTCACCGGACCATTCAA | [38] |
| OsEF1α | TTTCACTCTTGGTGTGAAGCAGAT | GACTTCCTTCACGATTTCATCGTAA | [37] |
| EcACCase | TAGAATTGCCGATCAATTTGTAGA | CTCCACTATGAGTTGAACATTTGC | a |
| EcCYP72A31 | AACCGACTCAAAACCGTGAC | CAGAACTGGCACTTCAACCA | |
| EcCYP81A12 | TCGGTAGTGGTGGAAAGAGC | AACGGAGTCTTGACGAGGTG | |
| EcGSTL2 | TTCCCTCGAGCACAATAACC | TGCTTCTTTGCAGCATCATC | |
| EcGSTL3 | GAGCGAGCTGCTTCTCAAGT | AAGGCCTCGTCGATGTACTG |
| aPOP | bGen | cEquation Parameters (SE) | dD50 | e CI 95% | |||
|---|---|---|---|---|---|---|---|
| A | b | R2 | p | ||||
| Visual injury | |||||||
| CHK | G0 | 0.103 (0.027) ** | 0.010 (0.001) ** | 0.84 | <0.01 | 10.3 | (6.3–17.5) |
| CHK | G1 | 0.090 (0.009) ** | 0.009 (0.002) ** | 0.93 | <0.01 | 8.2 | (5.3–12.0) |
| DRY | G1 | 0.874 (0.143) ** | 0.006 (0.001) ** | 0.76 | <0.01 | 62.4 | (36.5–96.8) |
| QPE | G1 | 0.593 (0.106) ** | 0.007 (0.009) ** | 0.74 | <0.01 | 45.6 | (25.3–73.2) |
| DRY × QPE | G1 | 0.200 (0.024) ** | 0.009 (0.003) ** | 0.92 | <0.01 | 18.2 | (12.6–22.5) |
| CHK | G2 | 0.195 (0.040) ** | 0.008 (0.005) ** | 0.80 | <0.01 | 16.2 | (8.8–25.0) |
| DRY | G2 | 0.816 (0.131) ** | 0.019 (0.002) ** | 0.80 | <0.01 | >200.0 | (138- > 200) |
| QPE | G2 | 0.398 (0.063) ** | 0.008 (0.006) ** | 0.81 | <0.01 | 33.2 | (20.8–52.5) |
| DRY × QPE | G2 | 0.326 (0.043) ** | 0.008 (0.004) ** | 0.86 | <0.01 | 27.2 | (18.4–37.4) |
| Shoot dry weight | |||||||
| CHK | G0 | 101.0 (5.10) ** | 0.51 (0.150) ** | 0.80 | <0.01 | 9.2 | (0–20.1) |
| CHK | G1 | 100.3 (3.96) ** | 0.541 (0.119) ** | 0.87 | <0.01 | 9.4 | (1.1–17.7) |
| DRY | G1 | 100.1 (5.38) ** | 0.750 (0.136) ** | 0.75 | <0.01 | 61.5 | (37.6–85.3) |
| QPE | G1 | 100.6 (7.30) ** | 0.905 (0.184) * | 0.66 | <0.01 | 43.6 | (24.0–63.0) |
| DRY × QPE | G1 | 100.3 (4.87) ** | 0.790 (0.165) ** | 0.83 | <0.01 | 12.2 | (6.0–24.4) |
| CHK | G2 | 100.4 (5.85) ** | 0.967 (0.199) ** | 0.78 | <0.01 | 23.6 | (12.6–34.4) |
| DRY | G2 | 99.9 (5.80) ** | 0.253 (0.145) ns | 0.40 | <0.01 | >200 | (>200) |
| QPE | G2 | 100.2 (7.09) ** | 0.684 (0.180) ** | 0.61 | <0.01 | 45.4 | (18.7–72.0) |
| DRY × QPE | G2 | 100.1 (6.29) ** | 0.527 (0.152) ** | 0.66 | <0.01 | 29.9 | (7.3–52.6) |
| b Collection Times (Hours after Applying) | ||||
|---|---|---|---|---|
| a POP | 24 | 48 | 96 | 144 |
| CHK | 0.206 c (0.18) | 0.794 (0.16) * | <LOQ | <LOQ |
| DRY | 0.050 (0.29) | d <LOQ | <LOQ | e N.D. |
| 48 | 96 | 192 | 336 | |
| CHK | <LOQ | <LOQ | <LOQ | <LOQ |
| DRY | 0.116 (0.16) | <LOQ | <LOQ | N.D |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fipke, M.V.; da Rosa Feijó, A.; Garcia, N.S.; Heck, T.; Viana, V.E.; Dayan, F.E.; Agostinetto, D.; Pinto Lamego, F.; Souza, G.M.; Camargo, E.R.; et al. Transgenerational Effect of Drought Stress and Sub-Lethal Doses of Quizalofop-p-ethyl: Decreasing Sensitivity to Herbicide and Biochemical Adjustment in Eragrostis plana. Agriculture 2022, 12, 396. https://doi.org/10.3390/agriculture12030396
Fipke MV, da Rosa Feijó A, Garcia NS, Heck T, Viana VE, Dayan FE, Agostinetto D, Pinto Lamego F, Souza GM, Camargo ER, et al. Transgenerational Effect of Drought Stress and Sub-Lethal Doses of Quizalofop-p-ethyl: Decreasing Sensitivity to Herbicide and Biochemical Adjustment in Eragrostis plana. Agriculture. 2022; 12(3):396. https://doi.org/10.3390/agriculture12030396
Chicago/Turabian StyleFipke, Marcus Vinícius, Anderson da Rosa Feijó, Natália Silva Garcia, Tamara Heck, Vívian Ebeling Viana, Franck Emmanuel Dayan, Dirceu Agostinetto, Fabiane Pinto Lamego, Gustavo Maia Souza, Edinalvo Rabaioli Camargo, and et al. 2022. "Transgenerational Effect of Drought Stress and Sub-Lethal Doses of Quizalofop-p-ethyl: Decreasing Sensitivity to Herbicide and Biochemical Adjustment in Eragrostis plana" Agriculture 12, no. 3: 396. https://doi.org/10.3390/agriculture12030396