Emodin Scavenging of Superoxide Radical Includes π–π Interaction. X-Ray Crystal Structure, Hydrodynamic Voltammetry and Theoretical Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Equipment
2.3. Superoxide Scavenging by Antioxidants
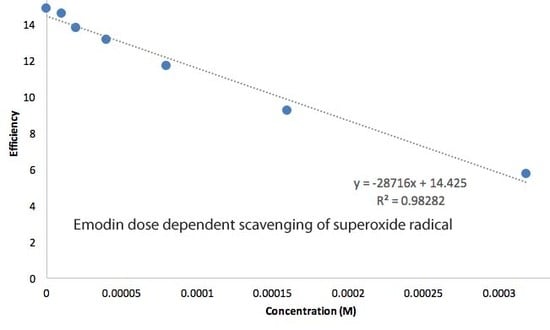
RRDE Study
2.4. Computational Study
2.5. Diffraction Study
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bucheli, P.; Gao, Q.; Redgwell, R.; Vidal, K.; Wang, J.; Zhang, W. Biomolecular and clinical aspects of chinese wolfberry. In Herbal Medicine: Biomolecular and Clinical Aspects, 2nd ed.; Benzie, I.F.F., Wachtel-Galor, S., Eds.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2011. [Google Scholar]
- Donno, D.; Beccaro, G.L.; Mellano, M.G.; Cerutti, A.K.; Bounous, G. Goji berry fruit (Lycium spp.): antioxidant compound fingerprint and bioactivity evaluation. J. Funct. Foods 2015, 18, 1070–1085. [Google Scholar] [CrossRef]
- Malik, E.M.; Müller, C.E. Anthraquinones as pharmacological tools and drugs. Med. Res. Rev. 2016, 36, 705–748. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-S.; Weng, S.-W.; Lin, M.-W.; Lu, C.-C.; Chiang, J.-H.; Yang, J.-S.; Chung, J.-G. Antitumor effects of emodin on LS1034 human colon cancer cells in vitro and in vivo: roles of apoptotic cell death and LS1034 tumor xenografts model. Food Chem. Toxicol. 2012, 50, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Rai, V. Role of emodin in chemosensltization of cancer. In Role of Nutraceuticals in Cancer Chemosensitization; Academic Press: Cambridge, MA, USA, 2018; Volume 2, pp. 241–257. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, T.; Huang, Y.; Chi, Q.; Shi, J.; Zhu, P.; Dong, N. Anthraquinone emodin inhibits tumor necrosis factor alpha-induced calcification of human aortic valve interstitial cells via the NF-κB pathway. Front. Pharmacol. 2018, 9, 1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Ni, Y.; Zhou, Q.; Liu, H.; Xiang, H.; Sui, H.; Shang, D. Emodin attenuates severe acute pancreatitis via antioxidant and anti-inflammatory activity. Inflammation 2019, 42, 2129–2138. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.L.; Yang, Y.; Liu, X.-L.; Xu, Q.-B. Emodin attenuates bleomycin-induced pulmonary fibrosis via anti-inflammatory and anti-oxidative activities in rats. Med. Sci. Monit. 2018, 24, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, P.Y.; Mak, D.H.F.; Poon, M.K.T.; Ko, K.M. In vivo antioxidant action of a lignan-enriched extract of Schisandra fruit and an anthraquinone-containing extract of Polygonum root in comparison with schisandrin B and emodin. Planta Med. 2002, 68, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Yen, G.-C.; Duh, P.-D.; Chuang, D.-Y. Antioxidant activity of anthraquinones and anthrone. Food Chem. 2000, 70, 437–441. [Google Scholar] [CrossRef]
- Mayrhofer, K.; Strmcnik, D.; Blizanac, B.; Stamenkovic, V.; Arenz, M.; Markovic, N. Measurement of oxygen reduction activities via the rotating disc electrode method: From Pt model surfaces to carbon-supported high surface area catalysts. Electrochim. Acta 2008, 53, 3181–3188. [Google Scholar] [CrossRef]
- Delley, B. From molecules to solids with the DMol3 approach. J. Chem. Phys. 2000, 113, 7756–7764. [Google Scholar] [CrossRef]
- Perdew, J.P.; Chevary, J.A.; Vosko, S.H.; Jackson, K.A.; Pederson, M.R.; Singh, D.J.; Fiolhais, C. Atoms, molecules, solids, and surfaces: Applications of the generalized gradient approximation for exchange and correlation. Phys. Rev. B Cond. Mat. Mater. Phys. 1992, 46, 6671–6687. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Wang, Y. Accurate and simple analytic representation of the electron-gas correlation energy. Phys. Rev. B 1992, 45, 13244–13249. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- From Crystal Structures to Patients. Available online: https://www.ccdc.cam.ac.uk (accessed on 25 February 2020).
- Zhu, J.-C.; Liang, Y.; Wang, H.-S.; Pan, Y.-M.; Zhang, Y. 1,3,8-Trihydr¬oxy-6-methyl-anthraquinone monohydrate. Acta Cryst. E 2007, 63, o233–o235. [Google Scholar] [CrossRef]
- Woińska, M.; Grabowsky, S.; Dominiak, P.M.; Woźniak, K.; Jayatilaka, D. Hydrogen atoms can be located accurately and precisely by x-ray crystallography. Sci. Adv. 2016, 2, e1600192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhang, Q.; He, H.; Wang, J.-R.; Mei, X. Vapor triggered fluorescent color changes among solvates of Emodin. J. Mater. Chem. C 2017, 5, 5970–5976. [Google Scholar] [CrossRef]
- Caruso, F.; Paumier, S.; Rossi, M. X-Ray crystal structure of embelin and its DFT scavenging of superoxide radical. J. Comput. Chem. 2018, 39, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Belli, S.; Rossi, M.; Molasky, N.; Middleton, L.; Caldwell, C.; Bartow-McKenney, C.; Duong, M.; Chiu, J.; Gibbs, E.; Caldwell, A.; et al. Effective and novel application of hydrodynamic voltammetry to the study of superoxide radical scavenging by natural phenolic antioxidants. Antioxidants 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013, 162750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emodin Reduction in Presence of O2. Available online: http://rpubs.com/SLBelli/553509 (accessed on 25 February 2020).















| Chemical formula | C15H12O6 | |
| Formula weight | 288.25 g/mol | |
| Temperature | 125(2) K | |
| Wavelength | 0.71073 Å | |
| Crystal size | 0.02 x 0.05 x 0.30 mm | |
| Crystal habit | clear orange-yellow needle | |
| Crystal system | monoclinic | |
| Space group | P 21/c | |
| Unit cell dimensions | a = 9.612(2) Å | α = 90° |
| b = 15.157(3) Å | β = 113.051(2)° | |
| c = 9.232(2) Å | γ = 90° | |
| Volume | 1237.6(5) Å3 | |
| Z | 4 | |
| Density (calculated) | 1.547 g/cm3 | |
| Absorption coefficient | 0.121 mm−1 | |
| F(000) | 600 | |
| Theta range for data collection | 2.30 to 28.28° | |
| Index ranges | −12<=h<=12, −20<=k<=20, −12<=l<=12 | |
| Reflections collected | 26402 | |
| Independent reflections | 3069 [R(int) = 0.1218] | |
| Coverage of independent reflections | 99.9% | |
| Absorption correction | Multi-Scan | |
| Max. and min. transmission | 0.9980 and 0.9650 | |
| Structure solution technique | direct methods | |
| Structure solution program | SHELXT 2014/5 [16] | |
| Refinement method | Full-matrix least-squares on F2 | |
| Refinement program | SHELXL-2016/6 [16] | |
| Function minimized | Σ w(Fo2 − Fc2)2 | |
| Data / restraints / parameters | 3069 / 0 / 238 | |
| Goodness-of-fit on F2 | 1.709 | |
| Δ/σmax | 0.012 | |
| Final R indices | 1643 data; I>2σ(I) | R1 = 0.0591, wR2 = 0.0548 |
| all data | R1 = 0.1390, wR2 = 0.0599 | |
| Weighting scheme | w=1/[σ2(Fo2)] where P=(Fo2+2Fc2)/3 | |
| Largest diff. peak and hole | 0.404 and -0.327 eÅ−3 | |
| R.M.S. deviation from mean | 0.070 eÅ−3 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, M.; Wen, K.; Caruso, F.; Belli, S. Emodin Scavenging of Superoxide Radical Includes π–π Interaction. X-Ray Crystal Structure, Hydrodynamic Voltammetry and Theoretical Studies. Antioxidants 2020, 9, 194. https://doi.org/10.3390/antiox9030194
Rossi M, Wen K, Caruso F, Belli S. Emodin Scavenging of Superoxide Radical Includes π–π Interaction. X-Ray Crystal Structure, Hydrodynamic Voltammetry and Theoretical Studies. Antioxidants. 2020; 9(3):194. https://doi.org/10.3390/antiox9030194
Chicago/Turabian StyleRossi, Miriam, Kelly Wen, Francesco Caruso, and Stuart Belli. 2020. "Emodin Scavenging of Superoxide Radical Includes π–π Interaction. X-Ray Crystal Structure, Hydrodynamic Voltammetry and Theoretical Studies" Antioxidants 9, no. 3: 194. https://doi.org/10.3390/antiox9030194