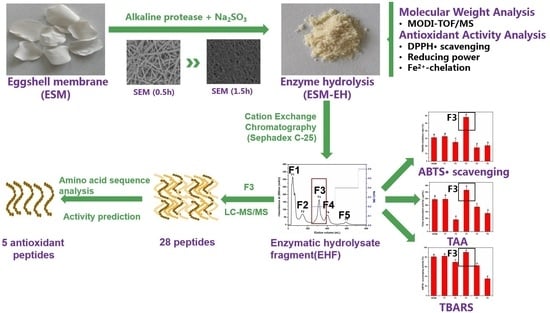
Separation and Identification of Highly Efficient Antioxidant Peptides from Eggshell Membrane
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation and Isolation of ESM-EH Fraction
2.3. Evaluation of the Antioxidant Activity of ESM-EH and EHF
2.4. Mass Spectrometry
2.4.1. MALDI-TOF/MS Analysis of ESM-EH
2.4.2. LC-MS/MS Analysis of EHF and Database Research
2.4.3. Performance Evaluation of Peptides
2.5. Statistical Analysis
3. Results and Discussion
3.1. Effect of Enzymatic Hydrolysis Parameters on the Antioxidant Activity of ESM-EH
3.2. Characterization of ESM-EH
3.2.1. Visual and Microscopical Analyses of ESM Membrane During the Enzymatic Hydrolysis Process
3.2.2. Analysis of Molecular Weight Distribution of ESM-EH by MALDI-TOF/MS
3.3. Analysis of the Correlation between Degree of Hydrolysis and Antioxidant Indices
3.4. Isolation and Identification of EHF in ESM-EH
3.4.1. Isolation and Characterization of EHF
3.4.2. Analysis of Amino Acid Sequence and Antioxidant Characteristics of F3
3.4.3. In Vitro Synthesis and Antioxidant Activity Verification of Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-Derived Bioactive Peptides on Inflammation and Oxidative Stress. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Kovacs-Nolan, J.; Jiang, B.; Tsao, R.; Mine, Y. Peptides derived from eggshell membrane improve antioxidant enzyme activity and glutathione synthesis against oxidative damage in Caco-2 cells. J. Funct. Foods 2014, 11, 571–580. [Google Scholar] [CrossRef]
- Moure, A.; Domínguez, H.; Parajó, J.C. Antioxidant properties of ultrafiltration-recovered soy protein fractions from industrial effluents and their hydrolysates. Process. Biochem. 2006, 41, 447–456. [Google Scholar] [CrossRef]
- Wu, S.; Sun, J.; Tong, Z.; Lan, X.; Zhao, Z.; Liao, D. Optimization of Hydrolysis Conditions for the Production of Angiotensin-I Converting Enzyme-Inhibitory Peptides and Isolation of a Novel Peptide from Lizard Fish (Saurida elongata) Muscle Protein Hydrolysate. Mar. Drugs 2012, 10, 1066–1080. [Google Scholar] [CrossRef] [Green Version]
- Shahidi, F.; Zhong, Y. Novel antioxidants in food quality preservation and health promotion. Eur. J. Lipid Sci. Technol. 2010, 112, 930–940. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Lee, B.H.; Oh, D.H. Current trends and perspectives of bioactive peptides. Crit. Rev. Food Sci. Nutr. 2018, 58, 2273–2284. [Google Scholar] [CrossRef]
- Stefanucci, A.; Mollica, A.; Macedonio, G.; Zengin, G.; Ahmed, A.A.; Novellino, E. Exogenous opioid peptides derived from food proteins and their possible uses as dietary supplements: A critical review. Food Rev. Int. 2016, 34, 70–86. [Google Scholar] [CrossRef]
- Esfandi, R.; Willmore, W.G.; Tsopmo, A. Peptidomic analysis of hydrolyzed oat bran proteins, and their in vitro antioxidant and metal chelating properties. Food Chem. 2019, 279, 49–57. [Google Scholar] [CrossRef]
- Zanutto-Elgui, M.R.; Vieira, J.C.S.; Prado, D.Z.D.; Buzalaf, M.A.R.; Padilha, P.D.M.; De Oliveira, D.E.; Fleuri, L.F. Production of milk peptides with antimicrobial and antioxidant properties through fungal proteases. Food Chem. 2019, 278, 823–831. [Google Scholar] [CrossRef]
- Martín-Del-Campo, S.T.; Martínez-Basilio, P.C.; Sepúlveda-Álvarez, J.C.; Gutiérrez-Melchor, S.E.; Galindo-Peña, K.D.; Lara-Domínguez, A.K.; Cardador-Martínez, A. Production of Antioxidant and ACEI Peptides from Cheese Whey Discarded from Mexican White Cheese Production. Antioxidants 2019, 8, 158. [Google Scholar] [CrossRef]
- Wedekind, K.J.; Ruff, K.J.; Atwell, C.A.; Evans, J.L.; Bendele, A.M. Beneficial effects of natural eggshell membrane (NEM) on multiple indices of arthritis in collagen-induced arthritic rats. Mod. Rheumatol. 2017, 27, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, Z.; Ahn, D.U.; Huang, X. Development of an antibacterial nanobiomaterial for wound-care based on the absorption of AgNPs on the eggshell membrane. Colloids Surfaces B Biointerfaces 2019, 183, 110449. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, M.; Ahn, D.U.; Huang, X. Preparation and characterization of novel eggshell membrane-chitosan blend films for potential wound-care dressing: From waste to medicinal products. Int. J. Boil. Macromol. 2019, 123, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Hogan, S.; Zhang, L.; Li, J.; Wang, H.; Zhou, K. Development of antioxidant rich peptides from milk protein by microbial proteases and analysis of their effects on lipid peroxidation in cooked beef. Food Chem. 2009, 117, 438–443. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, Y.; Ma, M.; Cai, Z.; Li, T. Chemiluminescence Evaluation of Antioxidant Activity and Prevention of DNA Damage Effect of Peptides Isolated from Soluble Eggshell Membrane Protein Hydrolysate. J. Agric. Food Chem. 2010, 58, 12137–12142. [Google Scholar] [CrossRef]
- Park, K.-M.; Yoo, J.-H.; Shin, Y.-J. Effects of Egg Shell Membrane Hydrolysates on Skin Whitening, Wound Healing, and UV-Protection. Food Sci. Anim. Resour. 2012, 32, 308–315. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.H.; Kim, J.K.; Yang, H.J.; Park, K.M. Effects of Egg Shell Membrane Hydrolysates on UVB-radiation-induced Wrinkle Formation in SKH-1 Hairless Mice. Food Sci. Anim. Resour. 2015, 35, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Shirai, K.; Kitamura, M.; Hattori, M. Soluble Egg Shell Membrane Protein as a Regulating Material for Collagen Matrix Reconstruction. Biosci. Biotechnol. Biochem. 1996, 60, 1299–1302. [Google Scholar] [CrossRef]
- Xu, S.; Shen, Y.; Chen, G.; Bean, S.; Li, Y. Antioxidant Characteristics and Identification of Peptides from Sorghum Kafirin Hydrolysates. J. Food Sci. 2019, 84, 2065–2076. [Google Scholar] [CrossRef]
- Ji, Y.; Chen, J.; Lv, J.; Li, Z.; Xing, L.; Ding, S. Extraction of keratin with ionic liquids from poultry feather. Sep. Purif. Technol. 2014, 132, 577–583. [Google Scholar] [CrossRef]
- Fan, J.; He, J.; Zhuang, Y.; Sun, L. Purification and Identification of Antioxidant Peptides from Enzymatic Hydrolysates of Tilapia (Oreochromis niloticus) Frame Protein. Molecules 2012, 17, 12836–12850. [Google Scholar] [CrossRef]
- Jahandideh, F.; Liu, P.; Wu, J. Purification and identification of adipogenic-differentiating peptides from egg white hydrolysate. Food Chem. 2018, 259, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duan, X.; Zhuang, Y. Purification and characterization of novel antioxidant peptides from enzymatic hydrolysates of tilapia (Oreochromis niloticus) skin gelatin. Peptides 2012, 38, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wattanasiritham, L.; Theerakulkait, C.; Wickramasekara, S.; Maier, C.S.; Stevens, J.F. Isolation and identification of antioxidant peptides from enzymatically hydrolyzed rice bran protein. Food Chem. 2016, 192, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, X.-Y.; Gan, R.-Y.; Zheng, J.; Li, Y.; Zhang, J.-J.; Xu, D.-P.; Li, H.-B. Optimization of Ultrasound-Assisted Extraction of Antioxidant Polyphenols from the Seed Coats of Red Sword Bean (Canavalia gladiate (Jacq.) DC.). Antioxidants 2019, 8. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Miao, M.; Jiang, B. Purification and characterisation of a new antioxidant peptide from chickpea (Cicer arietium L.) protein hydrolysates. Food Chem. 2011, 128, 28–33. [Google Scholar] [CrossRef]
- Fontoura, R.; Daroit, D.J.; Corrêa, A.P.F.; Moresco, K.S.; Santi, L.; Beys-Da-Silva, W.O.; Yates, J.R.; Moreira, J.C.F.; Brandelli, A. Characterization of a novel antioxidant peptide from feather keratin hydrolysates. New Biotechnol. 2019, 49, 71–76. [Google Scholar] [CrossRef]
- Yıldırım, A.; Mavi, A.; Kara, A.A.; Yildirim, A. Determination of Antioxidant and Antimicrobial Activities ofRumex crispusL. Extracts. J. Agric. Food Chem. 2001, 49, 4083–4089. [Google Scholar] [CrossRef]
- Huang, Y.; Ruan, G.; Qin, Z.; Li, H.; Zheng, Y. Antioxidant activity measurement and potential antioxidant peptides exploration from hydrolysates of novel continuous microwave-assisted enzymolysis of the Scomberomorus niphonius protein. Food Chem. 2017, 223, 89–95. [Google Scholar] [CrossRef]
- Le Grandois, J.; Guffond, D.; Hamon, E.; Marchioni, E.; Werner, D. Combined microplate-ABTS and HPLC-ABTS analysis of tomato and pepper extracts reveals synergetic and antagonist effects of their lipophilic antioxidative components. Food Chem. 2017, 223, 62–71. [Google Scholar] [CrossRef]
- Peptide Property Calculator. Available online: https://pepcalc.com/ (accessed on 3 January 2019).
- PeptideRanker. Available online: http://bioware.ucd.Ie/~compass/biowareweb/Server_pages/peptideranker.php (accessed on 5 January 2019).
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- BIOPEP. Available online: http://www.uwm.edu.pl/biochemia/index.php/pl/biopep (accessed on 5 January 2019).
- Kim, S.S.; Ahn, C.-B.; Moon, S.W.; Je, J.-Y. Purification and antioxidant activities of peptides from sea squirt (Halocynthia roretzi) protein hydrolysates using pepsin hydrolysis. Food Biosci. 2018, 25, 128–133. [Google Scholar] [CrossRef]
- Agrawal, H.; Joshi, R.; Gupta, M. Purification, identification and characterization of two novel antioxidant peptides from finger millet (Eleusine coracana) protein hydrolysate. Food Res. Int. 2019, 120, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, H.; Joshi, R.; Gupta, M. Isolation, purification and characterization of antioxidative peptide of pearl millet (Pennisetum glaucum) protein hydrolysate. Food Chem. 2016, 204, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Olagunju, A.I.; Omoba, O.S.; Enujiugha, V.N.; Alashi, A.M.; Aluko, R.E. Pigeon pea enzymatic protein hydrolysates and ultrafiltration peptide fractions as potential sources of antioxidant peptides: An in vitro study. LWT 2018, 97, 269–278. [Google Scholar] [CrossRef]
- Jakovetic, S.; Lukovic, N.; Jugovic, B.; Gvozdenovic, M.; Grbavcic, S.; Jovanovic, J.; Knezevic-Jugovic, Z. Production of Antioxidant Egg White Hydrolysates in a Continuous Stirred Tank Enzyme Reactor Coupled with Membrane Separation Unit. Food Bioprocess Technol. 2015, 8, 287–300. [Google Scholar] [CrossRef]
- Siddhanta, S. Separation and concentration of some platinum metal ions with a new chelating resin containing thiosemicarbazide as functional group. Talanta 1985, 32, 457–460. [Google Scholar] [CrossRef]
- Park, S.; Choi, K.S.; Lee, D.; Kim, D.; Lim, K.T.; Lee, K.-H.; Seonwoo, H.; Kim, J. Eggshell membrane: Review and impact on engineering. Biosyst Eng. 2016, 151, 446–463. [Google Scholar] [CrossRef]
- Yi, F.; Guo, Z.-X.; Zhang, L.-X.; Yu, J.; Li, Q. Soluble eggshell membrane protein: preparation, characterization and biocompatibility. Biomaterial 2004, 25, 4591–4599. [Google Scholar] [CrossRef]
- Katoh, K.; Tanabe, T.; Yamauchi, K. Novel approach to fabricate keratin sponge scaffolds with controlled pore size and porosity. Biomaterial 2004, 25, 4255–4262. [Google Scholar] [CrossRef]
- Soudham, V.P.; Alriksson, B.; Jönsson, L.J. Reducing agents improve enzymatic hydrolysis of cellulosic substrates in the presence of pretreatment liquid. J. Biotechnol. 2011, 155, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Makkar, S.; Liyanage, R.; Kannan, L.; Packialakshmi, B.; Lay, J.O.; Rath, N.C. Chicken Egg Shell Membrane Associated Proteins and Peptides. J. Agric. Food Chem. 2015, 63, 9888–9898. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Tian, W.; Li, H.; Cao, J.; Jiang, W. Improving antioxidant activities of whey protein hydrolysates obtained by thermal preheat treatment of pepsin, trypsin, alcalase and flavourzyme. Int. J. Food Sci. Technol. 2012, 47, 2045–2051. [Google Scholar] [CrossRef]
- Ren, Y.; Wu, H.; Li, X.; Lai, F.; Xiao, X. Purification and characterization of high antioxidant peptides from duck egg white protein hydrolysates. Biochem. Biophys. Res. Commun. 2014, 452, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kong, B.; Xiong, Y.L.; Xia, X. Antioxidant activity and functional properties of porcine plasma protein hydrolysate as influenced by the degree of hydrolysis. Food Chem. 2010, 118, 403–410. [Google Scholar] [CrossRef]
- Li, X.; Luo, Y.; Shen, H.; You, J. Antioxidant activities and functional properties of grass carp (Ctenopharyngodon idellus) protein hydrolysates. J. Sci. Food Agric. 2012, 92, 292–298. [Google Scholar] [CrossRef]
- Sun, Q.; Shen, H.; Luo, Y. Antioxidant activity of hydrolysates and peptide fractions derived from porcine hemoglobin. J. Food Sci. Technol. 2011, 48, 53–60. [Google Scholar] [CrossRef]
- Yang, R.; Li, X.; Lin, S.; Zhang, Z.; Chen, F. Identification of novel peptides from 3 to 10kDa pine nut (Pinus koraiensis) meal protein, with an exploration of the relationship between their antioxidant activities and secondary structure. Food Chem. 2017, 219, 311–320. [Google Scholar] [CrossRef]
- Pokora, M.; Zambrowicz, A.; Zabłocka, A.; Dąbrowska, A.; Szołtysik, M.; Babij, K.; Eckert, E.; Trziszka, T.; Chrzanowska, J. The use of serine protease from Yarrowia lipolytica yeast in the production of biopeptides from denatured egg white proteins. Acta Biochim. Pol. 2017, 64, 245–253. [Google Scholar] [CrossRef]
- Saiga, A.; Tanabe, S.; Nishimura, T. Antioxidant Activity of Peptides Obtained from Porcine Myofibrillar Proteins by Protease Treatment. J. Agric. Food Chem. 2003, 51, 3661–3667. [Google Scholar] [CrossRef]
- Nimalaratne, C.; Bandara, N.; Wu, J. Purification and characterization of antioxidant peptides from enzymatically hydrolyzed chicken egg white. Food Chem. 2015, 188, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Tanzadehpanah, H.; Asoodeh, A.; Chamani, J. An antioxidant peptide derived from Ostrich (Struthio camelus) egg white protein hydrolysates. Food Res. Int. 2012, 49, 105–111. [Google Scholar] [CrossRef]
- Li, Y.-W.; Li, B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: Key sequence positions and their amino acid properties. J. Theor. Boil. 2013, 318, 29–43. [Google Scholar] [CrossRef] [PubMed]







| Index | DPPH• Scavenging | Fe2+-Chelation | Reducing Power | TAA |
|---|---|---|---|---|
| DH | 0.465 | −0.529 * | 0.857 ** | 0.876 ** |
| DPPH• scavenging | - | −0.625 * | 0.827 ** | 0.573 * |
| Fe2+-chelation | - | - | −0.686 ** | –0.471 |
| Reducing power | - | - | - | 0.832 ** |
| No. | Peptide Amino Acid Sequence | Molecular Weight (Da) | Abundance | Water Solubility | Predicted Activity Score | Features of Anti-Oxidation | Gallus Gallus (Chicken) |
|---|---|---|---|---|---|---|---|
| 1 | KDQLTPSPR | 1040.6 | 1.72 × 108 | + | 0.33 | N-terminal (K). Intrinsic antioxidant activity (P L). Acid residues (D). | + |
| 2 | VEPKSPR | 811.5 | 3.47 × 107 | + | 0.19 | Intrinsic antioxidant activity (P V). Acid residues (E). | + |
| 3 | VEVYLPR | 874.5 | 7.82 × 107 | + | 0.27 | Intrinsic antioxidant activity (V L). The third amino acid to the left of the C-terminus (L). Acid residues (E). | + |
| 4 | RFRWSER | 1035.5 | 4.16 × 109 | + | 0.55 | Free radical scavenger (W). Acid residues (E). | + |
| 5 | MHSHIDMK | 997.4 | 1.64 × 108 | + | 0.43 | Free radical scavenger (H W M). C-terminal (K). Acid residues(D). | + |
| 6 | ESYHLPR | 900.4 | 7.26 × 107 | + | 0.52 | Free radical scavenger (Y H). Intrinsic antioxidant activity (P L). Acid residues (E). The third amino acid to the left of the C-terminus (L). | − |
| 7 | NVIDPPIYAR | 1156.6 | 5.46 × 107 | + | 0.42 | Free radical scavenger (Y). Acid residues (D). Intrinsic antioxidant activity (A P V). Repeat specific amino acids in adjacent positions (PP). | + |
| 8 | MFAEWQPR | 1063.5 | 8.35 × 106 | + | 0.69 | Free radical scavenger (M W F). Intrinsic antioxidant activity (P V). Acid residues (E). | + |
| 9 | MATWRRDGR | 1147.6 | 8.62 × 106 | + | 0.40 | Free radical scavenger (M W). Acid residues (D). Intrinsic antioxidant activity (A G). | + |
| 10 | MITLTELK | 947.5 | 6.70 × 106 | + | 0.09 | C-terminal (K). Free radical scavenger (M). Acid residues (E). | + |
| 11 | MEAAMGR | 764.3 | 3.18 × 105 | + | 0.35 | Free radical scavenger (M). Acid residues (E). | + |
| 12 | MQALSPR | 801.4 | 5.52 × 107 | + | 0.34 | Free radical scavenger (M). Intrinsic antioxidant activity (A P L). | − |
| 13 | RVWHKGR | 937.5 | 1.91 × 107 | + | 0.43 | Intrinsic antioxidant activity (V G). Free radical scavenger (H W). | − |
| 14 | MVGSKLPR | 886.5 | 1.11 × 107 | + | 0.36 | Intrinsic antioxidant activity (P V G L). Free radical scavenger (M). The third amino acid to the left of the C-terminus (L). | + |
| 15 | LLFAMTKPK | 1047.6 | 1.24 × 108 | + | 0.33 | Intrinsic antioxidant activity (A P L). C-terminal (K). N-terminal (L). Free radical scavenger(M). | + |
| 16 | MLKMLPFK | 1006.6 | 4.92 × 106 | + | 0.74 | C-terminal (K). Free radical scavenger (M F). Intrinsic antioxidant activity (P L). | + |
| 17 | ETLMGGPLR | 972.5 | 4.14 × 106 | + | 0.31 | Free radical scavenger (M). Intrinsic antioxidant activity (P L G). Acid residues (E). Repeat specific amino acids in adjacent positions (G G). | + |
| 18 | GTLHIQK | 795.5 | 1.39 × 108 | + | 0.14 | C-terminal (K). | + |
| 19 | GVLSPGR | 684.4 | 1.10 × 108 | + | 0.40 | Intrinsic antioxidant activity (G P V L). | + |
| 20 | SALPSPR | 726.4 | 2.97 × 108 | + | 0.64 | Intrinsic antioxidant activity (A P L). | − |
| 21 | PVSVAPR | 724.4 | 1.97 × 108 | + | 0.29 | Intrinsic antioxidant activity (A P V). | + |
| 22 | QQPVSPR | 810.4 | 5.95 × 107 | + | 0.37 | Intrinsic antioxidant activity (P V). | + |
| 23 | MMGGALQPR | 959.5 | 9.05 × 106 | − | 0.38 | Repeat specific amino acids in adjacent positions (GG). Intrinsic antioxidant activity (G A L P). | − |
| 24 | IPPVAVR | 750.5 | 2.56 × 107 | − | 0.29 | Repeat specific amino acids in adjacent positions (PP). N-terminal (I). | + |
| 25 | MALLLPWK | 970.6 | 2.15 × 107 | − | 0.81 | Free radical scavenger (M Y). C-terminal (K) | − |
| 26 | LGRVMYSMANCLLMMK | 1916.9 | 4.73 × 107 | − | 0.70 | Free radical scavenger (C M Y). Intrinsic antioxidant activity (G V A L). C-terminal (K). N-terminal (L). | + |
| 27 | MAHVQHLK | 962.5 | 2.69 × 107 | − | 0.23 | Free radical scavenger (M H). C-terminal (K), Intrinsic antioxidant activity (A L). | + |
| 28 | MISFCVMK | 1014.5 | 1.34 × 107 | − | 0.67 | C-terminal (K). Free radical scavenger (M F C). | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.-C.; Zhao, J.-Y.; Ahn, D.U.; Jin, Y.-G.; Huang, X. Separation and Identification of Highly Efficient Antioxidant Peptides from Eggshell Membrane. Antioxidants 2019, 8, 495. https://doi.org/10.3390/antiox8100495
Zhao Q-C, Zhao J-Y, Ahn DU, Jin Y-G, Huang X. Separation and Identification of Highly Efficient Antioxidant Peptides from Eggshell Membrane. Antioxidants. 2019; 8(10):495. https://doi.org/10.3390/antiox8100495
Chicago/Turabian StyleZhao, Qian-Cheng, Jie-Yuan Zhao, Dong Uk Ahn, Yong-Guo Jin, and Xi Huang. 2019. "Separation and Identification of Highly Efficient Antioxidant Peptides from Eggshell Membrane" Antioxidants 8, no. 10: 495. https://doi.org/10.3390/antiox8100495