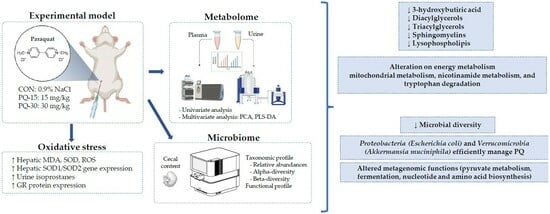
Exploring Metabolic and Gut Microbiome Responses to Paraquat Administration in Male Wistar Rats: Implications for Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. PQ Administration in Male Wistar Rats
2.2. Sample Collection
2.3. General Measurements for the Characterization of the Experimental Approach
2.3.1. Determinations in Plasma
2.3.2. Measurement of ROS in Liver Homogenates
2.3.3. RNA Extraction and qPCR
2.3.4. Protein Extraction and Western Blot Analysis
2.4. Plasma Metabolomics (GC-qTOF and UHPLC-qTOF)
2.5. Urine Metabolome (1H-NMR)
2.6. Shotgun Metagenomics Sequencing
2.7. Statistical Analysis
2.7.1. General Statistical Analysis
2.7.2. Metabolomic Data Analysis
2.7.3. Metagenomic Data Analysis
2.7.4. Multi-Omics Data Association with Isoprostanes (Gold Standard Biomarker)
2.7.5. Integration Data Analysis
2.7.6. Pathway Analysis
3. Results
3.1. Impact of PQ Administration on Physiological and Biochemical Responses
3.1.1. Body Changes and General Parameters in Rats
3.1.2. Oxidative and Inflammatory Profiling in Liver, Plasma and Urine
3.2. Exploring Plasma Metabolomic Alterations in Response to PQ Administration
3.3. Exploring Urine Metabolomic Alterations in Response to PQ Administration
3.4. Impact of PQ-Administration on Gut Microbiome
3.4.1. Taxonomic Impact of PQ-Administration
3.4.2. Functional Impact of PQ-Administration
3.5. Correlation of Isoprostane Levels with Multi-Omics Data Reveals Oxidative Injury Induced by PQ Administration
3.6. Multi-Omics Data Integration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dinis-Oliveira, R.J.; Duarte, J.A.; Sánchez-Navarro, A.; Remião, F.; Bastos, M.L.; Carvalho, F. Paraquat Poisonings: Mechanisms of Lung Toxicity, Clinical Features, and Treatment. Crit. Rev. Toxicol. 2008, 38, 13–71. [Google Scholar] [CrossRef] [PubMed]
- Seok, S.J.; Gil, H.W.; Jeong, D.S.; Yang, J.O.; Lee, E.Y.; Hong, S.Y. Paraquat Intoxication in Subjects Who Attempt Suicide: Why They Chose Paraquat. Korean J. Intern. Med. 2009, 24, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.R.; Chung, S.P.; You, J.S.; Cho, S.; Park, Y.; Chun, B.; Moon, J.; Kim, H.; Kim, Y.H.; Kim, H.J.; et al. Effects of Paraquat Ban on Herbicide Poisoning-Related Mortality. Yonsei Med. J. 2017, 58, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Dalvie, M.A.; White, N.; Raine, R.; Myers, J.E.; London, L.; Thompson, M.; Christiani, D.C. Long Term Respiratory Health Effects of the Herbicide, Paraquat, among Workers in the Western Cape. Occup. Environ. Med. 1999, 56, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Schenker, M.B.; Stoecklin, M.; Lee, K.; Lupercio, R.; Zeballos, R.J.; Enright, P.; Hennessy, T.; Beckett, L.A. Pulmonary Function and Exercise-Associated Changes with Chronic Low-Level Paraquat Exposure. Am. J. Respir. Crit. Care Med. 2004, 170, 773–779. [Google Scholar] [CrossRef]
- Tangamornsuksan, W.; Lohitnavy, O.; Sruamsiri, R.; Chaiyakunapruk, N.; Norman Scholfield, C.; Reisfeld, B.; Lohitnavy, M. Paraquat Exposure and Parkinson’s Disease: A Systematic Review and Meta-Analysis. Arch. Environ. Occup. Health 2019, 74, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Su, Y.; Lin, F.; Iqbal, M.; Mehmood, K.; Zhang, H.; Shi, D. Effect of Paraquat on Cytotoxicity Involved in Oxidative Stress and Inflammatory Reaction: A Review of Mechanisms and Ecological Implications. Ecotoxicol. Environ. Saf. 2021, 224, 112711. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, R.; Tiwari, R.R. The Herbicide Paraquat-Induced Molecular Mechanisms in the Development of Acute Lung Injury and Lung Fibrosis. Crit. Rev. Toxicol. 2021, 51, 36–64. [Google Scholar] [CrossRef]
- Al-Gubory, K.H.; Garrel, C.; Faure, P.; Sugino, N. Roles of Antioxidant Enzymes in Corpus Luteum Rescue from Reactive Oxygen Species-Induced Oxidative Stress. Reprod. Biomed. Online 2012, 25, 551–560. [Google Scholar] [CrossRef]
- Bouhifd, M.; Hartung, T.; Hogberg, H.T.; Kleensang, A.; Zhao, L. Review: Toxicometabolomics. J. Appl. Toxicol. 2013, 33, 1365–1383. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, J.; Zhang, M.; Wen, C.; Huang, X.; Sun, F.; Wang, S.; Hu, L.; Lin, G.; Wang, X. Serum Metabolomics in Rats after Acute Paraquat Poisoning. Biol. Pharm. Bull. 2015, 38, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zuo, Z.; Zhang, B.; Luo, H.; Song, B.; Zhou, Z.; Chang, X. Impacts of Early-Life Paraquat Exposure on Gut Microbiota and Body Weight in Adult Mice. Chemosphere 2022, 291, 133135. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, N.; Xiao, Y.; Deng, Y.; Zha, A.; Tan, B.; Wang, J.; Yin, Y.; Liao, P. Ellagic Acid Ameliorates Paraquat-Induced Liver Injury Associated with Improved Gut Microbial Profile. Environ. Pollut. 2022, 293, 118572. [Google Scholar] [CrossRef] [PubMed]
- Novaes, R.D.; Gonçalves, R.V.; Cupertino, M.C.; Santos, E.C.; Bigonha, S.M.; Fernandes, G.J.M.; Maldonado, I.R.S.C.; Natali, A.J. Acute Paraquat Exposure Determines Dose-Dependent Oxidative Injury of Multiple Organs and Metabolic Dysfunction in Rats: Impact on Exercise Tolerance. Int. J. Exp. Pathol. 2016, 97, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.F.; Fan, A.X.; Neuendorf, E.H.; Vergara, V.B.; Kalinich, J.F. Hydrophobic Sand Versus Metabolic Cages: A Comparison of Urine Collection Methods for Rats (Rattus norvegicus). J. Am. Assoc. Lab. Anim. Sci. 2018, 57, 51–57. [Google Scholar] [PubMed]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating Taxonomic, Functional, and Strain-Level Profiling of Diverse Microbial Communities with BioBakery 3. Elife 2021, 10, e65088. [Google Scholar] [CrossRef] [PubMed]
- Llorach-Asunción, R.; Jauregui, O.; Urpi-Sarda, M.; Andres-Lacueva, C. Methodological Aspects for Metabolome Visualization and Characterization: A Metabolomic Evaluation of the 24 h Evolution of Human Urine after Cocoa Powder Consumption. J. Pharm. Biomed. Anal. 2010, 51, 373–381. [Google Scholar] [CrossRef]
- Fujisaka, S.; Avila-Pacheco, J.; Soto, M.; Kostic, A.; Dreyfuss, J.M.; Pan, H.; Ussar, S.; Altindis, E.; Li, N.; Bry, L.; et al. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep. 2018, 22, 3072–3086. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Tenenhaus, M.; Tenenhaus, A.; Groenen, P.J.F. Regularized Generalized Canonical Correlation Analysis: A Framework for Sequential Multiblock Component Methods. Psychometrika 2017, 82, 737–777. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. MixOmics: An R Package for ’omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- González, I.; Cao, K.A.L.; Davis, M.J.; Déjean, S. Visualising Associations between Paired “omics” Data Sets. BioData Min. 2012, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.A.E.; El Morsy, E.M.; Ahmed, A.A.E. Protective Effects of Febuxostat against Paraquat-Induced Lung Toxicity in Rats: Impact on RAGE/PI3K/Akt Pathway and Downstream Inflammatory Cascades. Life Sci. 2019, 221, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Sharp, C.W.; Ottolenghi, A.; Posner, H.S. Correlation of Paraquat Toxicity with Tissue Concentrations and Weight Loss of the Rat. Toxicol. Appl. Pharmacol. 1972, 22, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Dicker, E.; Cederbaum, A.I. NADH-Dependent Generation of Reactive Oxygen Species by Microsomes in the Presence of Iron and Redox Cycling Agents. Biochem. Pharmacol. 1991, 42, 529–535. [Google Scholar] [CrossRef]
- Burk, R.F.; Lawrence, R.A.; Lane, J.M. Liver Necrosis and Lipid Peroxidation in the Rat as the Result of Paraquat and Diquat Administration. Effect of Selenium Deficiency. J. Clin. Investig. 1980, 65, 1024–1031. [Google Scholar] [CrossRef]
- Costa, M.D.; de Freitas, M.L.; Dalmolin, L.; Oliveira, L.P.; Fleck, M.A.; Pagliarini, P.; Acker, C.; Roman, S.S.; Brandão, R. Diphenyl Diselenide Prevents Hepatic Alterations Induced by Paraquat in Rats. Environ. Toxicol. Pharmacol. 2013, 36, 750–758. [Google Scholar] [CrossRef]
- Marí, M.; Morales, A.; Colell, A.; García-Ruiz, C.; Kaplowitz, N.; Fernández-Checa, J.C. Mitochondrial Glutathione: Features, Regulation and Role in Disease. Biochim. Biophys. Acta 2013, 1830, 3317–3328. [Google Scholar] [CrossRef]
- Tomita, M.; Katsuyama, H.; Okuyama, T.; Hidaka, K.; Minatogawa, Y. Changes in Gene Expression Level for Defense System Enzymes against Oxidative Stress and Glutathione Level in Rat Administered Paraquat. Int. J. Mol. Med. 2005, 15, 689–693. [Google Scholar] [CrossRef]
- Djukic, M.M.; Jovanovic, M.D.; Ninkovic, M.; Stevanovic, I.; Ilic, K.; Curcic, M.; Vekic, J. Protective Role of Glutathione Reductase in Paraquat Induced Neurotoxicity. Chem. Biol. Interact. 2012, 199, 74–86. [Google Scholar] [CrossRef]
- Liu, M.W.; Su, M.X.; Zhang, W.; Wang, Y.Q.; Chen, M.; Wang, L.; Qian, C.Y. Protective Effect of Xuebijing Injection on Paraquat-Induced Pulmonary Injury via down-Regulating the Expression of P38 MAPK in Rats. BMC Complement. Altern. Med. 2014, 14, 498. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.S.; Wang, I.K.; Liang, C.C.; Fu, J.F.; Hou, Y.C.; Chang, C.C.; Gu, P.W.; Tsai, K.F.; Weng, C.H.; Huang, W.H.; et al. Cytokine Changes in Fatal Cases of Paraquat Poisoning. Am. J. Transl. Res. 2021, 13, 11571. [Google Scholar] [PubMed]
- Hernandez-Baixauli, J.; Quesada-Vázquez, S.; Mariné-Casadó, R.; Cardoso, K.G.; Caimari, A.; Del Bas, J.M.; Escoté, X.; Baselga-Escudero, L. Detection of Early Disease Risk Factors Associated with Metabolic Syndrome: A New Era with the NMR Metabolomics Assessment. Nutrients 2020, 12, 806. [Google Scholar] [CrossRef] [PubMed]
- Frisard, M.; Ravussin, E. Energy Metabolism and Oxidative Stress: Impact on the Metabolic Syndrome and the Aging Process. Endocrine 2006, 29, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA Cycle Metabolites Control Physiology and Disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Tretter, L.; Adam-Vizi, V. Inhibition of Krebs Cycle Enzymes by Hydrogen Peroxide: A Key Role of [Alpha]-Ketoglutarate Dehydrogenase in Limiting NADH Production under Oxidative Stress. J. Neurosci. 2000, 20, 8972–8979. [Google Scholar] [CrossRef]
- Nulton-Persson, A.C.; Szweda, L.I. Modulation of Mitochondrial Function by Hydrogen Peroxide. J. Biol. Chem. 2001, 276, 23357–23361. [Google Scholar] [CrossRef]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl Coenzyme A: A Central Metabolite and Second Messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef]
- Rojas-Morales, P.; Pedraza-Chaverri, J.; Tapia, E. Ketone Bodies, Stress Response, and Redox Homeostasis. Redox Biol. 2020, 29, 101395. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-Dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Haces, M.L.; Hernández-Fonseca, K.; Medina-Campos, O.N.; Montiel, T.; Pedraza-Chaverri, J.; Massieu, L. Antioxidant Capacity Contributes to Protection of Ketone Bodies against Oxidative Damage Induced during Hypoglycemic Conditions. Exp. Neurol. 2008, 211, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Maalouf, M.; Sullivan, P.G.; Davis, L.; Kim, D.Y.; Rho, J.M. Ketones Inhibit Mitochondrial Production of Reactive Oxygen Species Production Following Glutamate Excitotoxicity by Increasing NADH Oxidation. Neuroscience 2007, 145, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Chiang, H.H.; Louw, M.; Susanto, A.; Chen, D. Nutrient Sensing and the Oxidative Stress Response. Trends Endocrinol. Metab. 2017, 28, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Jensen, N.J.; Wodschow, H.Z.; Nilsson, M.; Rungby, J. Effects of Ketone Bodies on Brain Metabolism and Function in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 8767. [Google Scholar] [CrossRef] [PubMed]
- Klosinski, L.P.; Yao, J.; Yin, F.; Fonteh, A.N.; Harrington, M.G.; Christensen, T.A.; Trushina, E.; Brinton, R.D. White Matter Lipids as a Ketogenic Fuel Supply in Aging Female Brain: Implications for Alzheimer’s Disease. EBioMedicine 2015, 2, 1888–1904. [Google Scholar] [CrossRef] [PubMed]
- Alessenko, A.V.; Shupik, M.A.; Gutner, U.A.; Bugrova, A.E.; Dudnik, L.B.; Shingarova, L.N.; Mikoyan, A.; Vanin, A.F. The Relation between Sphingomyelinase Activity, Lipid Peroxide Oxidation and NO-Releasing in Mice Liver and Brain. FEBS Lett. 2005, 579, 5571–5576. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Luberto, C.; Argraves, K.M. Enzymes of Sphingolipid Metabolism: From Modular to Integrative Signaling. Biochemistry 2001, 40, 4893–4903. [Google Scholar] [CrossRef]
- Zhou, X.; He, L.; Wu, C.; Zhang, Y.; Wu, X.; Yin, Y. Serine Alleviates Oxidative Stress via Supporting Glutathione Synthesis and Methionine Cycle in Mice. Mol. Nutr. Food Res. 2017, 61, 1700262. [Google Scholar] [CrossRef]
- Ducker, G.S.; Chen, L.; Morscher, R.J.; Ghergurovich, J.M.; Esposito, M.; Teng, X.; Kang, Y.; Rabinowitz, J.D. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016, 23, 1140–1153. [Google Scholar] [CrossRef]
- Palmnäs, M.S.A.; Kopciuk, K.A.; Shaykhutdinov, R.A.; Robson, P.J.; Mignault, D.; Rabasa-Lhoret, R.; Vogel, H.J.; Csizmadi, I. Serum Metabolomics of Activity Energy Expenditure and Its Relation to Metabolic Syndrome and Obesity. Sci. Rep. 2018, 8, 3308. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine Starvation Induces Stress and P53-Dependent Metabolic Remodelling in Cancer Cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, W.; Chen, C.; Yan, B.; Zhu, L.; Chen, X.; Peng, C. The Mechanisms of Lysophosphatidylcholine in the Development of Diseases. Life Sci. 2020, 247, 117443. [Google Scholar] [CrossRef] [PubMed]
- Giulivi, C.; Lavagno, C.C.; Lucesoli, F.; Bermüdez, M.J.É; Boveris, A. Lung Damage in Paraquat Poisoning and Hyperbaric Oxygen Exposure: Superoxide-Mediated Inhibition of Phospholipase A2. Free Radic. Biol. Med. 1995, 18, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.A.; Kim, J.A.; Park, M.H.; Jung, S.C.; Suh, S.H.; Pang, M.G.; Kim, Y.J. Lysophosphatidylcholine Induces Endothelial Cell Injury by Nitric Oxide Production through Oxidative Stress. J. Matern. Fetal Neonatal Med. 2009, 22, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Ostrakhovitch, E.A.; Tabibzadeh, S. Homocysteine in Chronic Kidney Disease. Adv. Clin. Chem. 2015, 72, 77–106. [Google Scholar] [CrossRef]
- Okabe, K.; Yaku, K.; Tobe, K.; Nakagawa, T. Implications of Altered NAD Metabolism in Metabolic Disorders. J. Biomed. Sci. 2019, 26, 34. [Google Scholar] [CrossRef]
- Topp, H.; Fusch, G.; Schöch, G.; Fusch, C. Noninvasive Markers of Oxidative DNA Stress, RNA Degradation and Protein Degradation Are Differentially Correlated with Resting Metabolic Rate and Energy Intake in Children and Adolescents. Pediatr. Res. 2008, 64, 246–250. [Google Scholar] [CrossRef]
- Miccheli, A.; Capuani, G.; Marini, F.; Tomassini, A.; Praticò, G.; Ceccarelli, S.; Gnani, D.; Baviera, G.; Alisi, A.; Putignani, L.; et al. Urinary (1)H-NMR-Based Metabolic Profiling of Children with NAFLD Undergoing VSL#3 Treatment. Int. J. Obes. 2015, 39, 1118–1125. [Google Scholar] [CrossRef]
- Dzarik, R.; Lajdova, I.; Spustova, V.; Opatrny, K., Jr. Pseudouridine Excretion in Healthy Subjects and Its Accumulation in Renal Failure. Nephron 1992, 61, 64–67. [Google Scholar] [CrossRef]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The Gut Microbiota: A Major Player in the Toxicity of Environmental Pollutants? NPJ Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef]
- Brown, J.B.; Langley, S.A.; Snijders, A.M.; Wan, K.H.; Morris, S.N.S.; Booth, B.W.; Fisher, W.W.; Hammonds, A.S.; Park, S.; Weiszmann, R.; et al. An Integrated Host-Microbiome Response to Atrazine Exposure Mediates Toxicity in Drosophila. Commun. Biol. 2021, 4, 1324. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The Resilience of the Intestinal Microbiota Influences Health and Disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.M.; Schellhorn, H.E. Regulators of Oxidative Stress Response Genes in Escherichia Coli and Their Functional Conservation in Bacteria. Arch. Biochem. Biophys. 2012, 525, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia Municiphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, Á.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The Abundance of Akkermansia Muciniphila and Its Relationship with Sulphated Colonic Mucins in Health and Ulcerative Colitis. Sci. Rep. 2019, 9, 15683. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Sunder, S.; Verma, S.R. Disease-Associated Dysbiosis and Potential Therapeutic Role of Akkermansia Muciniphila, a Mucus Degrading Bacteria of Gut Microbiome. Folia Microbiol. 2022, 67, 811–824. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A Purified Membrane Protein from Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Xie, S.; Zhang, R.; Li, Z.; Liu, C.; Xiang, W.; Lu, Q.; Chen, Y.; Yu, Q. Indispensable Role of Melatonin, a Scavenger of Reactive Oxygen Species (ROS), in the Protective Effect of Akkermansia Muciniphila in Cadmium-Induced Intestinal Mucosal Damage. Free Radic. Biol. Med. 2022, 193, 447–458. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Q.; Liu, M.; Zhang, X.; He, F.; Wang, G. Akkermansia Muciniphila Can Reduce the Damage of Gluco/Lipotoxicity, Oxidative Stress and Inflammation, and Normalize Intestine Microbiota in Streptozotocin-Induced Diabetic Rats. Pathog. Dis. 2018, 76, fty028. [Google Scholar] [CrossRef]
- Grajeda-lglesias, C.; Durand, S.; Daillére, R.; Iribarren, K.; Lemaitre, F.; Derosa, L.; Aprahamian, F.; Bossut, N.; Nirmalathasan, N.; Madeo, F.; et al. Oral Administration of Akkermansia Muciniphila Elevates Systemic Antiaging and Anticancer Metabolites. Aging 2021, 13, 6375–6405. [Google Scholar] [CrossRef]








| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Accession Number | Size (bp) |
|---|---|---|---|---|
| Cu/Zn SOD | GGTGGTCCACGAGAAACAAG | CAATCACACCACAAGCCAAG | NM_017050.1 | 98 |
| Mn SOD | AAGGAGCAAGGTCGCTTACA | ACACATCAATCCCCAGCAGT | NM_017051.2 | 94 |
| Catalase | GAATGGCTATGGCTCACACA | CAAGTTTTTGATGCCCTGGT | NM_012520.2 | 100 |
| GPx1 | TGCAATCAGTTCGGACATC | CACCTCGCACTTCTCAAACA | NM_030826.4 | 120 |
| PPIA | CCTCGAGCTGTTTGCAGACAA | AAGTCACCACCCTGGCACATG | NM_017101.1 | 138 |
| Corrected p-Value | Relative Abundance (%) | ||||||
|---|---|---|---|---|---|---|---|
| Specie | CON vs. PQ-15 vs. PQ-30 | PQ-15 vs. PQ-30 | CON vs. PQ-30 | CON vs. PQ-15 | CON | PQ-15 | PQ-30 |
| Muribaculaceae bacterium DSM 103720 | <0.01 *** | 0.02 ** | <0.01 *** | 1.00 | 30.40% | 34.43% | 0.03% |
| Akkermansia muciniphila | <0.01 *** | 0.02 ** | <0.01 *** | 1.00 | 2.30% | 2.75% | 19.31% |
| Muribaculum intestinale | <0.01 *** | 0.02 ** | <0.01 *** | 1.00 | 18.49% | 18.51% | - |
| Bifidobacterium pseudolongum | 0.01 ** | 0.02 ** | <0.01 *** | 1.00 | - | - | 0.15% |
| Anaerotruncus sp. G3 2012 | 0.01 ** | 0.02 ** | <0.01 *** | 1.00 | 1.87% | 1.44% | 0.04% |
| Escherichia coli | 0.01 ** | 0.02 ** | <0.01 *** | 1.00 | 10.79% | 6.09% | 76.00% |
| Oscillibacter sp. 1 3 | 0.01 ** | 0.02 ** | 0.08 * | 0.55 | 1.90% | 3.80% | 0.09% |
| Lactobacillus johnsonii | 0.02 ** | 0.03 ** | 0.01 ** | 1.00 | 0.07% | 0.01% | 0.16% |
| Bacteroides uniformis | 0.03 ** | 0.28 | 0.01 ** | 0.92 | 4.55% | 2.18% | 0.10% |
| Ruthenibacterium lactatiformans | 0.03 ** | 0.02 ** | 0.04 ** | 1.00 | 0.23% | 0.03% | 0.69% |
| Faecalibaculum rodentium | 0.04 ** | 0.05 * | 0.04 ** | 1.00 | - | - | 0.04% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez-Baixauli, J.; Chomiciute, G.; Tracey, H.; Mora, I.; Cortés-Espinar, A.J.; Ávila-Román, J.; Abasolo, N.; Palacios-Jordan, H.; Foguet-Romero, E.; Suñol, D.; et al. Exploring Metabolic and Gut Microbiome Responses to Paraquat Administration in Male Wistar Rats: Implications for Oxidative Stress. Antioxidants 2024, 13, 67. https://doi.org/10.3390/antiox13010067
Hernandez-Baixauli J, Chomiciute G, Tracey H, Mora I, Cortés-Espinar AJ, Ávila-Román J, Abasolo N, Palacios-Jordan H, Foguet-Romero E, Suñol D, et al. Exploring Metabolic and Gut Microbiome Responses to Paraquat Administration in Male Wistar Rats: Implications for Oxidative Stress. Antioxidants. 2024; 13(1):67. https://doi.org/10.3390/antiox13010067
Chicago/Turabian StyleHernandez-Baixauli, Julia, Gertruda Chomiciute, Harry Tracey, Ignasi Mora, Antonio J. Cortés-Espinar, Javier Ávila-Román, Nerea Abasolo, Hector Palacios-Jordan, Elisabet Foguet-Romero, David Suñol, and et al. 2024. "Exploring Metabolic and Gut Microbiome Responses to Paraquat Administration in Male Wistar Rats: Implications for Oxidative Stress" Antioxidants 13, no. 1: 67. https://doi.org/10.3390/antiox13010067