High Light Intensity Triggered Abscisic Acid Biosynthesis Mediates Anthocyanin Accumulation in Young Leaves of Tea Plant (Camellia sinensis)
Abstract
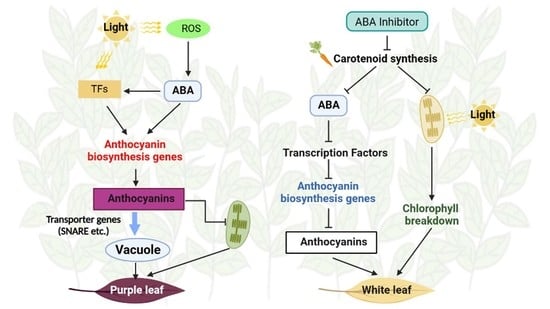
:1. Introduction
2. Materials and Methods
2.1. Monitoring Key Environmental Factors and Anthocyanin and ABA Contents in Young Leaves of ZFX 1 Cultivar
2.2. Effects of Shade Treatment with and without ABA on Pigment Contents in Young Leaves of ZFX1 and the Extracts’ Antioxidant Activities
2.3. Analysis of Pigments and Some Hormones in Young Leaves of Four Tea Cultivars and the Extracts’ Antioxidant Activities
2.4. Analysis of Pigment Contents in Young Leaves of ZFX1 Treated with ABA and ABA- Inhibitor and the Extracts’ Antioxidant Activities
2.5. Analyses of Pigments, Hormones, and Antioxidant Activities
2.6. Data Analysis of Aforementioned Experiments
2.7. Transcriptomic Analysis Leaves Treated with ABA, Flu or Water
2.8. Quantitative RT-PCR Analysis of Candidate Genes Involved in Anthocyanin Biosynthesis
3. Results
3.1. Environmental Factors Related to Anthocyanin and ABA Contents in Young Leaves of ZFX1
3.2. Anthocyanin Contents Affected by Shade Treatment and ABA Application
3.3. Anthocyanin and ABA Contents Were Substantially Higher in Purple Leaved Tea Cultivars
3.4. ABA and Flu Affected Anthocyanin Contents and Leaf Morphology of ZFX1
3.5. RNA–Seq and Functional Annotation
3.5.1. GO and KEGG Enrichment Analysis of DEGs
3.5.2. Exogenous Application of ABA and Flu Affected Anthocyanin Biosynthesis
3.5.3. DEGs Involved in Chlorophyll Metabolism and Light Signaling
3.6. Co-Expressed Gene Networks and Key Candidate Genes Related to Anthocyanin Biosynthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Zhang, X.; Zhou, L.; Wang, Y.; Fang, W.; Zhu, X. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 2018, 5, 7. [Google Scholar]
- Rashid, K.; Wachira, F.N.; Nyabuga, J.N.; Wanyonyi, B.; Murilla, G.; Isaac, A.O. Kenyan purple tea anthocyanins ability to cross the blood brain barrier and reinforce brain antioxidant capacity in mice. Nutr. Neurosci. 2014, 17, 178–185. [Google Scholar] [CrossRef]
- Zou, C.; Li, R.Y.; Chen, J.X.; Wang, F.; Gao, Y.; Fu, Y.Q.; Xu, Y.Q.; Yin, J.F. Zijuan tea-based kombucha: Physicochemical, sensorial, and antioxidant profile. Food Chem. 2021, 363, 130322. [Google Scholar] [CrossRef]
- Sun, B.; Zhu, Z.; Cao, P.; Chen, H.; Chen, C.; Zhou, X.; Mao, Y.; Lei, J.; Jiang, Y.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef]
- Tan, L.Q.; Yang, C.J.; Zhou, B.; Wang, L.B.; Zou, Y.; Chen, W.; Xia, T.; Tang, Q. Inheritance and quantitative trait loci analyses of the anthocyanins and catechins of Camellia sinensis cultivar ‘Ziyan’ with dark-purple leaves. Physiol. Plant. 2020, 170, 109–119. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Zheng, Y.; Gu, M.; Lin, X.; Wang, S.; Jin, S.; Ye, N. Comparison of Metabolome and Transcriptome of Flavonoid Biosynthesis Pathway in a Purple-Leaf Tea Germplasm Jinmingzao and a Green-Leaf Tea Germplasm Huangdan reveals Their Relationship with Genetic Mechanisms of Color Formation. Int. J. Mol. Sci. 2020, 21, 4167. [Google Scholar] [CrossRef]
- Zhou, Q.; Sun, W.; Lai, Z. Differential expression of genes in purple-shoot tea tender leaves and mature leaves during leaf growth. J. Sci. Food Agric. 2016, 96, 1982–1989. [Google Scholar] [CrossRef]
- Li, W.; Tan, L.; Zou, Y.; Tan, X.; Huang, J.; Chen, W.; Tang, Q. The Effects of Ultraviolet A/B Treatments on Anthocyanin Accumulation and Gene Expression in Dark-Purple Tea Cultivar ‘Ziyan’ (Camellia sinensis). Molecules 2020, 25, 354. [Google Scholar]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar]
- Feild, T.S.; Lee, D.W.; Holbrook, N.M. Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 2001, 127, 566–574. [Google Scholar]
- Bi, X.; Zhang, J.; Chen, C.; Zhang, D.; Li, P.; Ma, F. Anthocyanin contributes more to hydrogen peroxide scavenging than other phenolics in apple peel. Food Chem. 2014, 152, 205–209. [Google Scholar] [PubMed]
- Samkumar, A.; Jones, D.; Karppinen, K.; Dare, A.P.; Sipari, N.; Espley, R.V.; Martinussen, I.; Jaakola, L. Red and blue light treatments of ripening bilberry fruits reveal differences in signalling through abscisic acid-regulated anthocyanin biosynthesis. Plant Cell Environ. 2021, 44, 3227–3245. [Google Scholar] [PubMed]
- Malik, A.N.A.; Uddain, J.; Chin, C.K.; Chew, B.L.; Subramaniam, S. Elicitation of protocorm-like bodies (PLBs) of Dendrobium ‘Sabin Blue’ using methyl jasmonate, salicylic acid and melatonin for in vitro production of anthocyanin. Phytochem. Lett. 2021, 43, 60–64. [Google Scholar]
- Ferrara, G.; Mazzeo, A.; Matarrese, A.; Pacucci, C.; Gambacorta, G. Use of abscisic acid (S-ABA) and sucrose for improving color, anthocyanin content and antioxidant activity of ‘Crimson Seedless’ grape berries. Aust. J. Grape Wine Res. 2015, 21, 18–29. [Google Scholar]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar]
- Karppinen, K.; Tegelberg, P.; Haggman, H.; Jaakola, L. Abscisic Acid Regulates Anthocyanin Biosynthesis and Gene Expression Associated With Cell Wall Modification in Ripening Bilberry (Vaccinium myrtillus L.) Fruits. Front. Plant Sci. 2018, 9, 1259. [Google Scholar]
- Huang, J.; Zhao, X.; Chory, J. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019, 29, 4186–4199.e3. [Google Scholar]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef]
- Li, G.; Zhao, J.; Qin, B.; Yin, Y.; An, W.; Mu, Z.; Cao, Y. ABA mediates development-dependent anthocyanin biosynthesis and fruit coloration in Lycium plants. BMC Plant Biol. 2019, 19, 317. [Google Scholar] [CrossRef]
- Neff, M.M. Genetic Interactions between Phytochrome A, Phytochrome B, and Cryptochrome 1 during Arabidopsis Development. Plant Physiol. 1998, 118, 27–36. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and carotenoids: Measurement and characterization by UV-VIS spectroscopy. Current Protocols in Food Analytical Chemistry (CPFA). Curr. Protoc. Food Anal. Chem. 2001, 1, F4-3. [Google Scholar] [CrossRef]
- Wu, J.; Hettenhausen, C.; Meldau, S.; Baldwin, I.T. Herbivory Rapidly Activates MAPK Signaling in Attacked and Unattacked Leaf Regions but Not between Leaves of Nicotiana attenuata. Plant Cell 2007, 19, 1096–1122. [Google Scholar] [CrossRef]
- Bz, A.; Bm, A.; Cm, A.; Cx, A.; Jw, A.; Zw, A.; Dy, A.; Tao, X.B. Classification of Pu-erh ripened teas and their differences in chemical constituents and antioxidant capacity. LWT 2022, 153, 112370. [Google Scholar]
- Ma, B.; Wang, J.; Xu, C.; Wang, Z.; Yin, D.; Zhou, B.; Ma, C. Interrelation analysis between phenolic compounds and in vitro antioxidant activities in Pu-erh tea. LWT 2022, 158, 113117. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Walvoort, D.; Brus, D.J.; Gruijter, J. An R package for spatial coverage sampling and random sampling from compact geographical strata by k-means. Comput. Geosci. 2010, 36, 1261–1267. [Google Scholar] [CrossRef]
- Wang, P.; Chen, D.; Zheng, Y.; Jin, S.; Yang, J.; Ye, N. Identification and Expression Analyses of SBP-Box Genes Reveal Their Involvement in Abiotic Stress and Hormone Response in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 3404. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Singh, K.; Foley, R.C.; Onate-Sanchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Pourcel, L.; Irani, N.G.; Lu, Y.; Riedl, K.; Schwartz, S.; Grotewold, E. The formation of Anthocyanic Vacuolar Inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 2010, 3, 78–90. [Google Scholar] [CrossRef]
- Grotewold, E. The challenges of moving chemicals within and out of cells: Insights into the transport of plant natural products. Planta 2004, 219, 906–909. [Google Scholar] [CrossRef]
- Maritim, T.K.; Korir, R.K.; Nyabundi, K.W.; Wachira, F.N.; Kamunya, S.M.; Muoki, R.C. Molecular regulation of anthocyanin discoloration under water stress and high solar irradiance in pluckable shoots of purple tea cultivar. Planta 2021, 254, 85. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Gould, K.S.; McKelvie, J.; Markham, K.R. Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ. 2002, 25, 1261–1269. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.J.E.; Holcroft, D.M.; Jacobs, G. Anthocyanins in vegetative tissues: A proposed unified function in photoprotection. New Phytol. 2002, 155, 349–361. [Google Scholar] [CrossRef]
- Cho, D.; Shin, D.; Jeon, B.W.; Kwak, J.M. ROS-Mediated ABA Signaling. J. Plant Biol. 2009, 52, 102–113. [Google Scholar] [CrossRef]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klusener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Wang, X.-F.; Li, Y.-Y.; Song, L.-Q.; Zhao, L.-L.; You, C.-X.; Hao, Y.-J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS Induces Anthocyanin Production Via Late Biosynthetic Genes and Anthocyanin Deficiency Confers the Hypersensitivity to ROS-Generating Stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Liu, Z.; Chen, W.; Xu, H.; Feng, H. Anthocyanin Degrading and Chlorophyll Accumulation Lead to the Formation of Bicolor Leaf in Ornamental Kale. Int. J. Mol. Sci. 2019, 20, 603. [Google Scholar] [CrossRef] [PubMed]
- Nagamune, K.; Xiong, L.; Chini, E.; Sibley, L.D. Plants, endosymbionts and parasites: Abscisic acid and calcium signaling. Commun. Integr. Biol. 2008, 1, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.K.; Yu, K.J.; Liu, B.; Lan, Y.B.; Sun, R.Z.; Li, Q.; He, F.; Pan, Q.H.; Duan, C.Q.; Wang, J. Comparison of transcriptional expression patterns of carotenoid metabolism in ‘Cabernet Sauvignon’ grapes from two regions with distinct climate. J. Plant Physiol. 2017, 213, 75–86. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Cai, J.; Lv, L.; Zeng, X.; Zhang, F.; Chen, Y.; Tian, W.; Li, J.; Li, X.; Li, Y. Integrative Analysis of Metabolomics and Transcriptomics Reveals Molecular Mechanisms of Anthocyanin Metabolism in the Zikui Tea Plant (Camellia sinensis cv. Zikui). Int. J. Mol. Sci. 2022, 23, 4780. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.; Zhang, Y.; Ruan, L.; Li, H.; Wu, L.; Xu, L.; Zhang, C.; Zhou, X.; Cheng, H.; et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019, 97, 825–840. [Google Scholar] [CrossRef]
- Mei, Y.; Xie, H.; Liu, S.; Zhu, J.; Zhao, S.; Wei, C. Metabolites and Transcriptional Profiling Analysis Reveal the Molecular Mechanisms of the Anthocyanin Metabolism in the “Zijuan” Tea Plant (Camellia sinensis var. assamica). J. Agric. Food Chem. 2021, 69, 414–427. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Griesser, M.; Hoffmann, T.; Bellido, M.L.; Rosati, C.; Fink, B.; Kurtzer, R.; Aharoni, A.; Munoz-Blanco, J.; Schwab, W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008, 146, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Cutanda-Perez, M.C.; Ageorges, A.; Gomez, C.; Vialet, S.; Terrier, N.; Romieu, C.; Torregrosa, L. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 2009, 69, 633–648. [Google Scholar] [CrossRef]
- Given, N.K.; Venis, M.A.; Grierson, D. Phenylalanine Ammonia-Lyase Activity and Anthocyanin Synthesis in Ripening Strawberry Fruit. J. Plant Physiol. 1988, 133, 25–30. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Brandizz, F. News and views into the SNARE complexity in Arabidopsis. Front. Plant Sci. 2012, 3, 28. [Google Scholar] [CrossRef]
- Wang, L.; Yue, C.; Cao, H.; Zhou, Y.; Zeng, J.; Yang, Y.; Wang, X. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef]
- Yu, X.; Hu, S.; He, C.; Zhou, J.; Qu, F.; Ai, Z.; Chen, Y.; Ni, D. Chlorophyll Metabolism in Postharvest Tea (Camellia sinensis L.) Leaves: Variations in Color Values, Chlorophyll Derivatives, and Gene Expression Levels under Different Withering Treatments. J. Agric. Food Chem. 2019, 67, 10624–10636. [Google Scholar] [CrossRef]
- Li, F.F.; Wang, S.; Gu, S.C.; Cheng, D.W.; Gu, H.; Li, M.; Chen, J.Y.; Yang, Y.J. Effects of foliar application of Aba and PDJ on the coloration and quality of ‘Kyoho grape berry. J. Fruit Sci. 2020, 37, 362–370. [Google Scholar]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Lai, B.; Wang, D.; Li, J.; Chen, L.; Qin, Y.; Wang, H.; Qin, Y.; Hu, G.; Zhao, J. Three LcABFs are Involved in the Regulation of Chlorophyll Degradation and Anthocyanin Biosynthesis during Fruit Ripening in Litchi chinensis. Plant Cell Physiol. 2019, 60, 448–461. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, X.; Lin, N.; Yu, S.; Fernie, A.R.; Zhao, J. CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator-repressor network. Hortic. Res. 2021, 8, 110. [Google Scholar] [CrossRef]
- Tattini, M.; Sebastiani, F.; Brunetti, C.; Fini, A.; Torre, S.; Gori, A.; Centritto, M.; Ferrini, F.; Landi, M.; Guidi, L. Dissecting molecular and physiological response mechanisms to high solar radiation in cyanic and acyanic leaves: A case study on red and green basil. J. Exp. Bot. 2017, 68, 2425–2437. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Avramenko, T.V.; Tsitsiashvili, G.S. Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: Focus on anthocyanins. Crit. Rev. Biotechnol. 2017, 37, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Espley, R.V.; Stevenson, D.; Cooney, J.; Datson, P.M.; Saiz, A.; Atkinson, R.G.; Hellens, R.P.; Allan, A.C. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2011, 65, 106–118. [Google Scholar] [CrossRef]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Li, X.J.; Bing, H.; Qin, Y.H.; Huang, X.M.; Wang, H.C.; Hu, G.B. LcMYB1 Is a Key Determinant of Differential Anthocyanin Accumulation among Genotypes, Tissues, Developmental Phases and ABA and Light Stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Bowen, B.; Chandler, V.L. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Du, L.-N.; Liu, R.; Hu, B.; Su, W.-B.; Qin, Y.-H.; Zhao, J.-T.; Wang, H.-C.; Hu, G.-B. Two LcbHLH Transcription Factors Interacting with LcMYB1 in Regulating Late Structural Genes of Anthocyanin Biosynthesis in Nicotiana and Litchi chinensis during Anthocyanin Accumulation. Front. Plant Sci. 2016, 7, 166. [Google Scholar] [CrossRef]
- Yadav, V.; Mallappa, C.; Gangappa, S.N.; Bhatia, S.; Chattopadhyay, S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 2005, 17, 1953–1966. [Google Scholar] [CrossRef] [Green Version]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Sun, Y.; Li, J.; Zhou, Z.; Deng, X.; Wang, Z.; Wu, S.; Lin, L.; Huang, Y.; Zeng, W.; et al. High Light Intensity Triggered Abscisic Acid Biosynthesis Mediates Anthocyanin Accumulation in Young Leaves of Tea Plant (Camellia sinensis). Antioxidants 2023, 12, 392. https://doi.org/10.3390/antiox12020392
Gao C, Sun Y, Li J, Zhou Z, Deng X, Wang Z, Wu S, Lin L, Huang Y, Zeng W, et al. High Light Intensity Triggered Abscisic Acid Biosynthesis Mediates Anthocyanin Accumulation in Young Leaves of Tea Plant (Camellia sinensis). Antioxidants. 2023; 12(2):392. https://doi.org/10.3390/antiox12020392
Chicago/Turabian StyleGao, Chenxi, Yue Sun, Jing Li, Zhe Zhou, Xuming Deng, Zhihui Wang, Shaoling Wu, Lin Lin, Yan Huang, Wen Zeng, and et al. 2023. "High Light Intensity Triggered Abscisic Acid Biosynthesis Mediates Anthocyanin Accumulation in Young Leaves of Tea Plant (Camellia sinensis)" Antioxidants 12, no. 2: 392. https://doi.org/10.3390/antiox12020392