A Machine Learning Algorithm for Quantitatively Diagnosing Oxidative Stress Risks in Healthy Adult Individuals Based on Health Space Methodology: A Proof-of-Concept Study Using Korean Cross-Sectional Cohort Data
Abstract
:1. Introduction
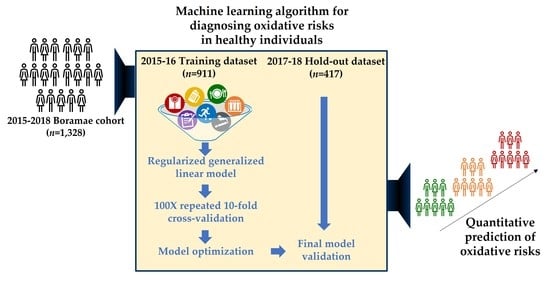
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.3. MDA Analysis
2.4. Development and Validation of ML Algorithm
2.5. Statistics
3. Results
3.1. Characteristics of the Reference Groups
3.2. Developing an ML Algorithm for Discriminating Oxidative Stress Risks in the Healthy Population
3.3. Internal and External Validation of the Best Performing Model
3.4. Application of the Best Performing Model to All Subjects in the Whole Dataset for Testing Discrimination Power
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonomini, F.; Rodella, L.F.; Rezzani, R. Metabolic syndrome, aging and involvement of oxidative stress. Aging Dis. 2015, 6, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Campbell, A.; Solaimani, P. Oxidative and inflammatory pathways in age-related chronic disease processes. In Inflammation, Aging, and Oxidative Stress; Springer: Berlin/Heidelberg, Germany, 2016; pp. 95–106. [Google Scholar]
- Ruiz-Núñez, B.; Pruimboom, L.; Dijck-Brouwer, D.A.; Muskiet, F.A. Lifestyle and nutritional imbalances associated with Western diseases: Causes and consequences of chronic systemic low-grade inflammation in an evolutionary context. J. Nutr. Biochem. 2013, 24, 1183–1201. [Google Scholar] [CrossRef] [Green Version]
- Hanson, M.A.; Gluckman, P.D. Developmental origins of health and disease--global public health implications. Best Pract. Res. Clin. Obstet. Gynaecol. 2015, 29, 24–31. [Google Scholar] [CrossRef]
- Bloomer, R.J. Decreased blood antioxidant capacity and increased lipid peroxidation in young cigarette smokers compared to nonsmokers: Impact of dietary intake. Nutr. J. 2007, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, F.; Mikkelsen, B.B.; Nielsen, J.B.; Andersen, H.R.; Grandjean, P. Plasma malondialdehyde as biomarker for oxidative stress: Reference interval and effects of life-style factors. Clin. Chem. 1997, 43, 1209–1214. [Google Scholar] [CrossRef] [Green Version]
- Cherubini, A.; Ruggiero, C.; Polidori, M.C.; Mecocci, P. Potential markers of oxidative stress in stroke. Free Radic. Biol. Med. 2005, 39, 841–852. [Google Scholar] [CrossRef]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity: Implications for metabolic syndrome, diabetes, hypertension, dyslipidemia, atherosclerosis, and cancer. Obes. Res. Clin. Pract. 2013, 7, e330–e341. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Br. J. Surg. 2015, 102, 148–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Elmasry, G.; Kamruzzaman, M.; Sun, D.W.; Allen, P. Principles and applications of hyperspectral imaging in quality evaluation of agro-food products: A review. Crit. Rev. Food Sci. Nutr. 2012, 52, 999–1023. [Google Scholar] [CrossRef]
- Bouwman, J.; Vogels, J.T.; Wopereis, S.; Rubingh, C.M.; Bijlsma, S.; Ommen, B. Visualization and identification of health space, based on personalized molecular phenotype and treatment response to relevant underlying biological processes. BMC Med. Genom. 2012, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- Murdoch, T.B.; Detsky, A.S. The inevitable application of big data to health care. JAMA 2013, 309, 1351–1352. [Google Scholar] [CrossRef]
- Dilsizian, S.E.; Siegel, E.L. Artificial intelligence in medicine and cardiac imaging: Harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr. Cardiol. Rep. 2014, 16, 441. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Yang, Y.J.; Yang, Y.K.; Oh, S.Y.; Hong, Y.C.; Lee, E.K.; Kwon, O. Diet quality scores and oxidative stress in Korean adults. Eur. J. Clin. Nutr. 2011, 65, 1271–1278. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.Y.; Yang, Y.J.; Kim, B.S.; Kang, J.H. Validity and reliability of Korean version of International Physical Activity Questionnaire (IPAQ) short form. J. Korean Acad. Fam. Med. 2007, 28, 532–541. [Google Scholar]
- Agarwal, R.; Chase, S.D. Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 775, 121–126. [Google Scholar] [CrossRef]
- Khoschsorur, G.; Winklhofer-Roob, B.; Rabl, H.; Auer, T.; Peng, Z.; Schaur, R. Evaluation of a sensitive HPLC method for the determination of malondialdehyde, and application of the method to different biological materials. Chromatographia 2000, 52, 181–184. [Google Scholar] [CrossRef]
- Arsova-Sarafinovska, Z.; Aydin, A.; Sayal, A.; Eken, A.; Erdem, O.; Savaşer, A.; Erten, K.; Özgök, Y.; Dimovski, A. Rapid and simple determination of plasma and erythrocyte MDA levels in prostate cancer patients by a validated HPLC method. J. Liq. Chromatogr. Relat. Technol. 2007, 30, 2435–2444. [Google Scholar] [CrossRef]
- Cipierre, C.; Haÿs, S.; Maucort-Boulch, D.; Steghens, J.-P.; Picaud, J.-C. Malondialdehyde adduct to hemoglobin: A new marker of oxidative stress suitable for full-term and preterm neonates. Oxidative Med. Cell. Longev. 2013, 2013, 1–6. [Google Scholar] [CrossRef]
- Maritim, A.C.; Sanders, R.A.; Watkins, J.B., 3rd. Diabetes, oxidative stress, and antioxidants: A review. J. Biochem. Mol. Toxicol. 2003, 17, 24–38. [Google Scholar] [CrossRef]
- Natarajan, S.K.; Thomas, S.; Ramamoorthy, P.; Basivireddy, J.; Pulimood, A.B.; Ramachandran, A.; Balasubramanian, K.A. Oxidative stress in the development of liver cirrhosis: A comparison of two different experimental models. J. Gastroenterol. Hepatol. 2006, 21, 947–957. [Google Scholar] [CrossRef]
- Kohler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021, 49, D1207–D1217. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef]
- Cohen, Z.D.; Kim, T.T.; Van, H.L.; Dekker, J.J.M.; Driessen, E. A demonstration of a multi-method variable selection approach for treatment selection: Recommending cognitive-behavioral versus psychodynamic therapy for mild to moderate adult depression. Psychother. Res. 2020, 30, 137–150. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1. [Google Scholar] [CrossRef] [Green Version]
- Steyerberg, E.W.; Bleeker, S.E.; Moll, H.A.; Grobbee, D.E.; Moons, K.G. Internal and external validation of predictive models: A simulation study of bias and precision in small samples. J. Clin. Epidemiol. 2003, 56, 441–447. [Google Scholar] [CrossRef]
- Melkumova, L.E.; Shatskikh, S.Y. Comparing Ridge and LASSO estimators for data analysis. Procedia Eng. 2017, 201, 746–755. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Bolboacă, S.D. Medical diagnostic tests: A review of test anatomy, phases, and statistical treatment of data. Comput. Math. Methods Med. 2019, 2019, 1–22. [Google Scholar] [CrossRef] [Green Version]
- D’Agostino, R.B.; Nam, B.-H. Evaluation of the Performance of Survival Analysis Models: Discrimination and Calibration Measures. In Advances in Survival Analysis; Elsevier: Amsterdam, The Netherlands, 2003; pp. 1–25. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. [Google Scholar]
- Moons, K.G.; Altman, D.G.; Vergouwe, Y.; Royston, P. Prognosis and prognostic research: Application and impact of prognostic models in clinical practice. BMJ 2009, 338, b606. [Google Scholar] [CrossRef]
- Block, G.; Dietrich, M.; Norkus, E.P.; Morrow, J.D.; Hudes, M.; Caan, B.; Packer, L. Factors associated with oxidative stress in human populations. Am. J. Epidemiol. 2002, 156, 274–285. [Google Scholar] [CrossRef] [Green Version]
- Yamada, J.; Tomiyama, H.; Yambe, M.; Koji, Y.; Motobe, K.; Shiina, K.; Yamamoto, Y.; Yamashina, A. Elevated serum levels of alanine aminotransferase and gamma glutamyltransferase are markers of inflammation and oxidative stress independent of the metabolic syndrome. Atherosclerosis 2006, 189, 198–205. [Google Scholar] [CrossRef]
- Waggiallah, H.; Alzohairy, M. The effect of oxidative stress on human red cells glutathione peroxidase, glutathione reductase level, and prevalence of anemia among diabetics. N. Am. J. Med. Sci. 2011, 3, 344–347. [Google Scholar] [CrossRef]
- Widmer, C.C.; Pereira, C.P.; Gehrig, P.; Vallelian, F.; Schoedon, G.; Buehler, P.W.; Schaer, D.J. Hemoglobin can attenuate hydrogen peroxide-induced oxidative stress by acting as an antioxidative peroxidase. Antioxid. Redox Signal. 2010, 12, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Roehrs, M.; Conte, L.; da Silva, D.T.; Duarte, T.; Maurer, L.H.; de Carvalho, J.A.M.; Moresco, R.N.; Somacal, S.; Emanuelli, T. Annatto carotenoids attenuate oxidative stress and inflammatory response after high-calorie meal in healthy subjects. Food Res. Int. 2017, 100, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rosales, B.L.; Tamez-Pena, J.G.; Nicolini, H.; Moreno-Treviño, M.G.; Trevino, V. Improving predictive models for Alzheimer’s disease using GWAS data by incorporating misclassified samples modeling. PLoS ONE 2020, 15, e0232103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, E.; Carroll, J.A.; Smith, H.E.; Scott, D.; Cassell, J.A. Extracting information from the text of electronic medical records to improve case detection: A systematic review. J. Am. Med. Inform. Assoc. 2016, 23, 1007–1015. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.Y.; Cheng, C.W.; Kaddi, C.D.; Venugopalan, J.; Hoffman, R.; Wang, M.D. -Omic and Electronic Health Record Big Data Analytics for Precision Medicine. IEEE Trans. Biomed. Eng. 2017, 64, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R.J.A.o.s. Additive logistic regression: A statistical view of boosting (with discussion and a rejoinder by the authors). Ann. Statist. 2000, 28, 337–407. [Google Scholar] [CrossRef]
- Dhillon, V.; Thomas, P.; Fenech, M. Effect of common polymorphisms in folate uptake and metabolism genes on frequency of micronucleated lymphocytes in a South Australian cohort. Mutat. Res. 2009, 665, 1–6. [Google Scholar] [CrossRef]
- Winham, S.J.; Slater, A.J.; Motsinger-Reif, A.A. A comparison of internal validation techniques for multifactor dimensionality reduction. BMC Bioinform. 2010, 11, 394. [Google Scholar] [CrossRef] [Green Version]
- Steyerberg, E.W.; Harrell, F.E.; Borsboom, G.J.; Eijkemans, M.J.; Vergouwe, Y.; Habbema, J.D. Internal validation of predictive models: Efficiency of some procedures for logistic regression analysis. J. Clin. Epidemiol. 2001, 54, 774–781. [Google Scholar] [CrossRef]
- Carter, J.V.; Pan, J.; Rai, S.N.; Galandiuk, S. ROC-ing along: Evaluation and interpretation of receiver operating characteristic curves. Surgery 2016, 159, 1638–1645. [Google Scholar] [CrossRef]
- Park, S.; Yoo, H.J.; Jee, S.H.; Lee, J.H.; Kim, M. Weighting approaches for a genetic risk score and an oxidative stress score for predicting the incidence of obesity. Diabetes Metab. Res. Rev. 2019, e3230. [Google Scholar] [CrossRef]
- Vassalle, C.; Pratali, L.; Boni, C.; Mercuri, A.; Ndreu, R. An oxidative stress score as a combined measure of the pro-oxidant and anti-oxidant counterparts in patients with coronary artery disease. Clin. Biochem. 2008, 41, 1162–1167. [Google Scholar] [CrossRef]
- Groenwold, R.H.; White, I.R.; Donders, A.R.; Carpenter, J.R.; Altman, D.G.; Moons, K.G. Missing covariate data in clinical research: When and when not to use the missing-indicator method for analysis. CMAJ 2012, 184, 1265–1269. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z. Multiple imputation with multivariate imputation by chained equation (MICE) package. Ann. Transl. Med. 2016, 4, 30. [Google Scholar] [CrossRef]





| Features | 2015–2016 Training Set (n = 610) | p-Value | 2017–2018 Hold-Out Set (n = 274) | p-Value | ||
|---|---|---|---|---|---|---|
| Healthy Controls (n = 248) | Oxidative Disease Cases (n = 362) | Healthy Controls (n = 131) | Oxidative Disease Cases (n = 143) | |||
| General characteristics (10) | ||||||
| Age (years) | 42.7 ± 10.4 | 52.5 ± 10.2 | <0.0001 | 38.3 ± 10.0 | 52.4 ± 10.1 | <0.0001 |
| Sex (males; n, %) | 101 (40.7) | 264 (72.9) | <0.0001 | 47 (35.9) | 102 (71.3) | <0.0001 |
| Current smoker (n, %) | 23 (9.3) | 97 (26.8) | <0.0001 | 5 (3.8) | 15 (10.5) | 0.034 |
| Smoking duration (y) | 4.5 ± 9.0 | 11.7 ± 13.1 | <0.0001 | 3.1 ± 6.8 | 9.3 ± 12.6 | <0.0001 |
| Smoking pack-year | 3.3 ± 7.6 | 9.5 ± 12.7 | <0.0001 | 2.4 ± 6.4 | 9.3 ± 15.7 | <0.0001 |
| Current drinker (n, %) | 145 (58.5) | 224 (61.9) | 0.397 | 78 (59.5) | 82 (57.3) | 0.712 |
| RFS | 21.6 ± 8.3 | 24.7 ± 8.7 | <0.0001 | 20.5 ± 8.7 | 22.5 ± 9.0 | 0.069 |
| Physical activity (m/d) | 128.5 ± 120.1 | 133.9 ± 118.9 | 0.581 | 119.2 ± 113.8 | 81.0 ± 107.0 | 0.005 |
| BMI (kg/m2) | 21.5 ± 2.2 | 25.6 ± 4.1 | <0.0001 | 21.2 ± 2.2 | 25.4 ± 3.7 | <0.0001 |
| Body fat (%) | 25.5 (6.4) | 28.6 (6.9) | <0.0001 | 26.0 (6.4) | 29.3 (6.5) | <0.0001 |
| Biochemical characteristics (14) | ||||||
| Albumin (g/dL) | 4.3 ± 0.2 | 4.4 ± 0.2 | 0.089 | 4.4 ± 0.2 | 4.5 ± 0.2 | 0.079 |
| AP (IU/L) | 62.5 ± 17.8 | 72.6 ± 18.5 | <0.0001 | 62.3 ± 17.2 | 70.1 ± 17.8 | <0.0001 |
| Bilirubin (mg/dL) | 1.2 ± 0.5 | 1.1 ± 0.4 | 0.129 | 1.1 ± 0.4 | 1.1 ± 0.4 | 0.237 |
| BUN (mg/dL) | 12.8 ± 3.5 | 13.9 ± 3.4 | <0.0001 | 11.6 ± 2.8 | 13.8 ± 3.3 | <0.0001 |
| Creatinine (µmol/L) | 68.2 ± 14.1 | 77.0 ± 15.5 | <0.0001 | 66.1 ± 12.6 | 77.6 ± 15.8 | <0.0001 |
| CRP (mg/dL) | 0.1 ± 0.2 | 0.2 ± 0.3 | <0.0001 | 0.1 ± 0.3 | 0.2 ± 0.4 | <0.0001 |
| GT (IU/L) | 18.4 ± 15.0 | 38.3 ± 38.2 | <0.0001 | 17.7 ± 13.7 | 39.7 ± 36.0 | <0.0001 |
| GOT (IU/L) | 22.6 ± 6.8 | 30.5 ± 18.4 | <0.0001 | 26.7 ± 56.0 | 31.2 ± 17.0 | 0.372 |
| GPT (IU/L) | 18.9 ± 12.0 | 35.5 ± 32.4 | <0.0001 | 18.6 ± 12.3 | 34.2 ± 24.2 | <0.0001 |
| Glycosylated Hb (%) | 5.4 ± 0.3 | 6.0 ± 0.9 | <0.0001 | 5.3 ± 0.3 | 5.9 ± 0.9 | <0.0001 |
| LDL-C (mmol/L) | 3.0 ± 0.7 | 3.1 ± 0.9 | 0.223 | 3.0 ± 0.7 | 3.2 ± 1.1 | 0.028 |
| TC (mmol/L) | 5.0 ± 0.8 | 5.1 ± 1.1 | 0.293 | 5.0 ± 0.8 | 5.2 ± 1.1 | 0.089 |
| Total protein (g/dL) | 7.1 ± 0.4 | 7.2 ± 0.4 | 0.283 | 7.2 ± 0.3 | 7.3 ± 0.4 | 0.081 |
| Uric acid (mg/dL) | 4.8 ± 1.2 | 5.6 ± 1.4 | <0.0001 | 4.7 ± 1.1 | 5.6 ± 1.3 | <0.0001 |
| Complete blood count data (16) | ||||||
| Basophils (%) | 0.4 ± 0.3 | 0.4 ± 0.3 | 0.504 | 0.5 ± 0.3 | 0.4 ± 0.3 | 0.273 |
| Eosinophils (%) | 2.6 ± 2.2 | 2.8 ± 2.2 | 0.273 | 2.7 ± 2.0 | 2.9 ± 2.5 | 0.419 |
| ESR (mm/h) | 9.0 ± 8.2 | 9.6 ± 8.4 | 0.380 | 9.4 ± 11.9 | 11.8 ± 11.1 | 0.090 |
| Hematocrit (%) | 41.4 ± 4.2 | 44.2 ± 3.8 | <0.0001 | 42.7 ± 3.7 | 44.9 ± 3.7 | <0.0001 |
| Hemoglobin (g/dL) | 13.9 ± 1.6 | 15.0 ± 1.5 | <0.0001 | 14.2 ± 1.5 | 15.1 ± 1.4 | <0.0001 |
| Lymphocytes (%) | 35.3 ± 8.8 | 35.4 ± 8.1 | 0.883 | 36.4 ± 7.9 | 36.3 ± 7.6 | 0.893 |
| MCH (pg) | 30.3 ± 1.9 | 30.8 ± 1.5 | <0.0001 | 30.0 ± 1.9 | 30.4 ± 1.3 | 0.094 |
| MCHC (g/dL) | 33.5 ± 1.0 | 34.0 ± 0.9 | <0.0001 | 33.2 ± 0.9 | 33.5 ± 0.9 | 0.002 |
| MCV (fL) | 90.4 ± 4.4 | 90.8 ± 3.9 | 0.209 | 90.6 ± 4.7 | 90.7 ± 3.4 | 0.830 |
| Monocytes (%) | 5.7 ± 1.5 | 5.6 ± 1.4 | 0.424 | 5.8 ± 1.4 | 5.9 ± 1.5 | 0.709 |
| Neutrophils (%) | 56.0 ± 9.7 | 55.8 ± 8.6 | 0.775 | 54.7 ± 8.4 | 54.6 ± 8.5 | 0.905 |
| Platelets (×103/µL) | 249.1 ± 51.9 | 247.9 ± 53.4 | 0.781 | 258.5 ± 54.0 | 258.4 ± 86.8 | 0.995 |
| PDW (%) | 11.8 ± 1.6 | 11.9 ± 1.5 | 0.412 | 11.8 ± 1.5 | 12.2 ± 1.6 | 0.016 |
| RBC count (×106/µL) | 4.6 ± 0.4 | 4.9 ± 0.5 | <0.0001 | 4.7 ± 0.4 | 5.0 ± 0.4 | <0.0001 |
| RDW (%) | 13.1 ± 1.0 | 12.9 ± 0.6 | 0.014 | 13.1 ± 1.1 | 12.9 ± 0.6 | 0.188 |
| WBC count (×103/µL) | 5.2 ± 1.3 | 6.2 ± 1.7 | <0.0001 | 5.2 ± 1.3 | 6.1 ± 1.8 | <0.0001 |
| High-performance liquid chromatography analysis data (3) | ||||||
| Plasma MDA (mM) | 4.4 ± 2.4 | 5.0 ± 2.5 | 0.009 | 4.3 ± 2.2 | 4.8 ± 2.0 | 0.083 |
| Erythrocyte MDA (mM) | 6.1 ± 6.2 | 6.3 ± 6.9 | 0.601 | 8.8 ± 8.6 | 9.4 ± 7.7 | 0.564 |
| Urine MDA (mM) | 2.9 ± 1.8 | 2.7 ± 1.5 | 0.380 | 2.7 ± 1.7 | 2.5 ± 1.3 | 0.207 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Kim, Y.; Hwang, J.; van den Broek, T.J.; Oh, B.; Kim, J.Y.; Wopereis, S.; Bouwman, J.; Kwon, O. A Machine Learning Algorithm for Quantitatively Diagnosing Oxidative Stress Risks in Healthy Adult Individuals Based on Health Space Methodology: A Proof-of-Concept Study Using Korean Cross-Sectional Cohort Data. Antioxidants 2021, 10, 1132. https://doi.org/10.3390/antiox10071132
Kim Y, Kim Y, Hwang J, van den Broek TJ, Oh B, Kim JY, Wopereis S, Bouwman J, Kwon O. A Machine Learning Algorithm for Quantitatively Diagnosing Oxidative Stress Risks in Healthy Adult Individuals Based on Health Space Methodology: A Proof-of-Concept Study Using Korean Cross-Sectional Cohort Data. Antioxidants. 2021; 10(7):1132. https://doi.org/10.3390/antiox10071132
Chicago/Turabian StyleKim, Youjin, Yunsoo Kim, Jiyoung Hwang, Tim J. van den Broek, Bumjo Oh, Ji Yeon Kim, Suzan Wopereis, Jildau Bouwman, and Oran Kwon. 2021. "A Machine Learning Algorithm for Quantitatively Diagnosing Oxidative Stress Risks in Healthy Adult Individuals Based on Health Space Methodology: A Proof-of-Concept Study Using Korean Cross-Sectional Cohort Data" Antioxidants 10, no. 7: 1132. https://doi.org/10.3390/antiox10071132