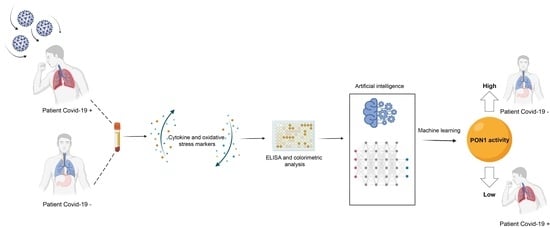
Clinical Performance of Paraoxonase-1-Related Variables and Novel Markers of Inflammation in Coronavirus Disease-19. A Machine Learning Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Biochemical Analyses
2.3. Development of the Predictive Models by Machine Learning
2.4. Statistical Analyses
3. Results
3.1. Relationships between PON1-Related Variables and Novel Inflammation Markers with the Clinical Characteristics of the Study Groups
3.2. Machine Learning Identified Serum PON1 Activity as the Best Analytical Parameter to Discriminate between COVID-19 Positive and Negative Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lartigue, L.; Faustin, B. Mitochondria: Metabolic regulators of innate immune responses to pathogens and cell stress. Int. J. Biochem. Cell. Biol. 2013, 45, 2052–2056. [Google Scholar] [CrossRef] [PubMed]
- Monlun, M.; Hyernard, C.; Blanco, P.; Lartigue, L.; Faustin, B. Mitochondria as molecular platforms integrating multiple innate immune signalings. J. Mol. Biol. 2017, 429, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.S.; Suh, H.W.; Kim, S.J.; Jo, E.K. Mitochondrial control of innate immunity and inflammation. Immune Netw. 2017, 17, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Camps, J.; Rodríguez-Gallego, E.; García-Heredia, A.; Triguero, I.; Riera-Borrull, M.; Hernández-Aguilera, A.; Luciano-Mateo, F.; Fernández-Arroyo, S.; Joven, J. Paraoxonases and chemokine (C-C motif) ligand-2 in noncommunicable diseases. Adv. Clin. Chem. 2014, 63, 247–308. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Iftimie, S.; García-Heredia, A.; Castro, A.; Joven, J. Paraoxonases and infectious diseases. Clin. Biochem. 2017, 50, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Marsillach, J.; Joven, J. The paraoxonases: Role in human diseases and methodological difficulties in measurement. Crit. Rev. Clin. Lab. Sci. 2009, 46, 83–106. [Google Scholar] [CrossRef] [PubMed]
- Parra, S.; Alonso-Villaverde, C.; Coll, B.; Ferré, N.; Marsillach, J.; Aragonès, G.; Mackness, M.; Mackness, B.; Masana, L.; Joven, J.; et al. Serum paraoxonase-1 activity and concentration are influenced by human immunodeficiency virus infection. Atherosclerosis 2007, 194, 175–181. [Google Scholar] [CrossRef]
- Farid, A.S.; Horii, Y. Modulation of paraoxonases during infectious diseases and its potential impact on atherosclerosis. Lipids Health Dis. 2012, 11, 92. [Google Scholar] [CrossRef] [Green Version]
- Sans, T.; Rull, A.; Luna, J.; Mackness, B.; Mackness, M.; Joven, J.; Ibañez, M.; Pariente, R.; Rodriguez, M.; Ortin, X.; et al. Monocyte chemoattractant protein-1 and paraoxonase-1 and 3 levels in patients with sepsis treated in an intensive care unit: A preliminary report. Clin. Chem. Lab. Med. 2012, 50, 1409–1415. [Google Scholar] [CrossRef] [PubMed]
- Iftimie, S.; García-Heredia, A.; Pujol, I.; Ballester, F.; Fort-Gallifa, I.; Simó, J.M.; Joven, J.; Camps, J.; Castro, A. Preliminary study on serum paraoxonase-1 status and chemokine (C-C motif) ligand 2 in hospitalized elderly patients with catheter-associated asymptomatic bacteriuria. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Iftimie, S.; García-Heredia, A.; Pujol, I.; Ballester, F.; Fort-Gallifa, I.; Simó, J.M.; Joven, J.; Castro, A.; Camps, J. A preliminary study of paraoxonase-1 in infected patients with an indwelling central venous catheter. Clin. Biochem. 2016, 49, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Fort-Gallifa, I.; Hernández-Aguilera, A.; García-Heredia, A.; Cabré, N.; Luciano-Mateo, F.; Simó, J.M.; Martín-Paredero, V.; Camps, J.; Joven, J. Galectin-3 in peripheral artery disease. Relationships with markers of oxidative stress and inflammation. Int. J. Mol. Sci. 2017, 18, 973. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Ouellet, M.; St-Pierre, C.; Tremblay, M.J. Glycans, galectins, and HIV-1 infection. Ann. N. Y. Acad. Sci. 2012, 1253, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Fort-Gallifa, I.; García-Heredia, A.; Hernández-Aguilera, A.; Simó, J.M.; Sepúlveda, J.; Martín-Paredero, V.; Camps, J.; Joven, J. Biochemical indices of oxidative stress and inflammation in the evaluation of peripheral artery disease. Free Radic. Biol. Med. 2016, 97, 568–576. [Google Scholar] [CrossRef] [PubMed]
- McCabe, W.R.; Jackson, G.G. Gram-negative bacteremia. I. Etiology and ecology. Arch. Intern. Med. 1962, 110, 847–864. [Google Scholar] [CrossRef]
- Berkman, L.F.; Leo-Summers, L.; Horwitz, R.I. Emotional support and survival after myocardial infarction. A prospective, population-based study of the elderly. Ann. Intern. Med. 1992, 117, 1003–1009. [Google Scholar] [CrossRef] [Green Version]
- Gaita, L.; Manzi, B.; Sacco, R.; Lintas, C.; Altieri, L.; Lombardi, F.; Pawlowski, T.L.; Redman, M.; Craig, D.W.; Huentelman, M.J.; et al. Decreased serum arylesterase activity in autism spectrum disorders. Psychiatry Res. 2010, 180, 105–113. [Google Scholar] [CrossRef]
- Zweig, M.H.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef]
- Kuris, E.O.; Veeramani, A.; McDonald, C.L.; DiSilvestro, K.J.; Zhang, A.S.; Cohen, E.M.; Daniels, A.H. Predicting readmission after anterior, posterior, and posterior interbody lumbar spinal fusion: A neural network machine learning approach. World Neurosurg. 2021, S1878-8750(21)00307-7. [Google Scholar] [CrossRef]
- Abraham, A.; Pedregosa, F.; Eickenberg, M.; Gervais, P.; Mueller, A.; Kossaifi, J.; Gramfort, A.; Thirion, B.; Varoquaux, G. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 2014, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, J.D.; Guerrero, M.D. An R package for computing Canadian assessment of physical literacy (CAPL) scores and interpretations from raw data. PLoS ONE 2021, 16, e0243841. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Kim, J.R.; Lee, I.C.; Kwon, H.J. Native high-density lipoproteins (HDL) with higher paraoxonase exerts a potent antiviral effect against SARS-CoV-2 (COVID-19), while glycated HDL lost the antiviral activity. Antioxidants 2021, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Cava, C.; Bertoli, G.; Castiglioni, I. In silico discovery of candidate drugs against Covid-19. Viruses 2020, 12, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, B.T.; Borchardt, R.T. Adenosine dialdehyde: A potent inhibitor of vaccinia virus multiplication in mouse L929 cells. Mol. Pharmacol. 1987, 31, 485–492. [Google Scholar] [PubMed]
- Hilser, J.R.; Han, Y.; Biswas, S.; Gukasyan, J.; Cai, Z.; Zhu, R.; Tang, W.H.W.; Deb, A.; Lusis, A.J.; Hartiala, J.A.; et al. Association of serum HDL-cholesterol and apolipoprotein A1 levels with risk of severe SARS-CoV-2 infection. J. Lipid Res. 2021, 62, 100061. [Google Scholar] [CrossRef] [PubMed]
- Masana, L.; Correig, E.; Ibarretxe, D.; Anoro, E.; Arroyo, J.A.; Jericó, C.; Guerrero, C.; Miret, M.; Näf, S.; Pardo, A.; et al. STACOV-XULA research group. Low HDL and high triglycerides predict COVID-19 severity. Sci. Rep. 2021, 11, 7217. [Google Scholar] [CrossRef]
- Begue, F.; Tanaka, S.; Mouktadi, Z.; Rondeau, P.; Veeren, B.; Diotel, N.; Tran-Dinh, A.; Robert, T.; Vélia, E.; Mavingui, P.; et al. Altered high-density lipoprotein composition and functions during severe COVID-19. Sci. Rep. 2021, 11, 2291. [Google Scholar] [CrossRef]
- Ferré, N.; Feliu, A.; García-Heredia, A.; Marsillach, J.; París, N.; Zaragoza-Jordana, M.; Mackness, B.; Mackness, M.; Escribano, J.; Closa-Monasterolo, R.; et al. Impaired paraoxonase-1 status in obese children. Relationships with insulin resistance and metabolic syndrome. Clin. Biochem. 2013, 46, 1830–1836. [Google Scholar] [CrossRef]
- Martinelli, N.; García-Heredia, A.; Roca, H.; Aranda, N.; Arija, V.; Mackness, B.; Mackness, M.; Busti, F.; Aragonès, G.; Pedro-Botet, J.; et al. Paraoxonase-1 status in patients with hereditary hemochromatosis. J. Lipid Res. 2013, 54, 1484–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsillach, J.; Aragonès, G.; Mackness, B.; Mackness, M.; Rull, A.; Beltrán-Debón, R.; Pedro-Botet, J.; Alonso-Villaverde, C.; Joven, J.; Camps, J. Decreased paraoxonase-1 activity is associated with alterations of high-density lipoprotein particles in chronic liver impairment. Lipids Health Dis. 2010, 9, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsillach, J.; Camps, J.; Ferré, N.; Beltran, R.; Rull, A.; Mackness, B.; Mackness, M.; Joven, J. Paraoxonase-1 is related to inflammation, fibrosis and PPAR delta in experimental liver disease. BMC Gastroenterol. 2009, 9, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferré, N.; Marsillach, J.; Camps, J.; Mackness, B.; Mackness, M.; Riu, F.; Coll, B.; Tous, M.; Joven, J. Paraoxonase-1 is associated with oxidative stress, fibrosis and FAS expression in chronic liver diseases. J. Hepatol. 2006, 45, 51–59. [Google Scholar] [CrossRef]
- Mackness, B.; Hine, D.; Liu, Y.; Mastorikou, M.; Mackness, M. Paraoxonase-1 inhibits oxidised LDL-induced MCP-1 production by endothelial cells. Biochem. Biophys. Res. Commun. 2004, 318, 680–683. [Google Scholar] [CrossRef]
- Sozer, V.; Himmetoglu, S.; Korkmaz, G.G.; Kaya, S.; Aydin, S.; Yumuk, V.; Hatemi, H.; Uzun, H. Paraoxonase, oxidized low density lipoprotein, monocyte chemoattractant protein-1 and adhesion molecules are associated with macrovascular complications in patients with type 2 diabetes mellitus. Minerva Med. 2014, 105, 237-44. [Google Scholar] [PubMed]
- Rull, A.; García, R.; Fernández-Sender, L.; Beltrán-Debón, R.; Aragonès, G.; Alegret, J.M.; Alonso-Villaverde, C.; Mackness, B.; Mackness, M.; Camps, J.; et al. The role of combined assessment of defense against oxidative stress and inflammation in the evaluation of peripheral arterial disease. Curr. Mol. Med. 2011, 11, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Haljasmägi, L.; Salumets, A.; Rumm, A.P.; Jürgenson, M.; Krassohhina, E.; Remm, A.; Sein, H.; Kareinen, L.; Vapalahti, O.; Sironen, T.; et al. Longitudinal proteomic profiling reveals increased early inflammation and sustained apoptosis proteins in severe COVID-19. Sci. Rep. 2020, 10, 20533. [Google Scholar] [CrossRef] [PubMed]
- Jøntvedt Jørgensen, M.; Holter, J.C.; Christensen, E.E.; Schjalm, C.; Tonby, K.; Pischke, S.E.; Jenum, S.; Skeie, L.G.; Nur, S.; Lind, A.; et al. Increased interleukin-6 and macrophage chemoattractant protein-1 are associated with respiratory failure in COVID-19. Sci. Rep. 2020, 10, 21697. [Google Scholar] [CrossRef]
- Li, S.; Jiang, L.; Li, X.; Lin, F.; Wang, Y.; Li, B.; Jiang, T.; An, W.; Liu, S.; Liu, H.; et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight 2020, 5, e138070. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Guo, Y.; Zhu, M.; Wei, Y.; Li, G.; Du, B.; Wang, Y. Higher expression of monocyte chemotactic protein 1 in mild COVID-19 patients might be correlated with inhibition of Type I IFN signaling. Virol. J. 2021, 18, 12. [Google Scholar] [CrossRef]
- Zhang, F.; Mears, J.R.; Shakib, L.; Beynor, J.I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium; Donlin, L.T.; Raychaudhuri, S. IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med. 2021, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chan, J.F.; Wang, Y.; Yuen, T.T.; Chai, Y.; Hou, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020, 71, 1400–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Lu, L.; Cao, W.; Li, T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020, 9, 727–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, W.; Zhou, Q.; Yao, S.; Deng, Q.; Wang, T.; Wu, Q. Mechanical ventilation induces an inflammatory response in preinjured lungs in late phase of sepsis. Oxid. Med. Cell. Longev. 2015, 2015, 364020. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Lin, C.Y.; Chang, M.R.; Urbina, A.N.; Assavalapsakul, W.; Thitithanyanont, A.; Chen, Y.H.; Liu, F.T.; Wang, S.F. The role of galectins in virus infection-A systemic literature review. J. Microbiol. Immunol. Infect. 2020, 53, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Caniglia, J.L.; Asuthkar, S.; Tsung, A.J.; Guda, M.R.; Velpula, K.K. Immunopathology of galectin-3: An increasingly promising target in COVID-19. F1000Research 2020, 9, 1078. [Google Scholar] [CrossRef] [PubMed]
- Kazancioglu, S.; Yilmaz, F.M.; Bastug, A.; Ozbay, B.O.; Aydos, O.; Yücel, Ç.; Bodur, H.; Yilmaz, G. Assessment of galectin-1, galectin-3, and PGE2 levels in patients with COVID-19. Jpn. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef] [PubMed]
- Caniglia, J.L.; Guda, M.R.; Asuthkar, S.; Tsung, A.J.; Velpula, K.K. A potential role for galectin-3 inhibitors in the treatment of COVID-19. PeerJ 2020, 8, e9392. [Google Scholar] [CrossRef] [PubMed]






| Healthy Subjects n = 50 | COVID-19 Negative Patients n = 45 | COVID-19 Positive Patients n = 126 | ||
|---|---|---|---|---|
| Demographic variables | ||||
| Sex, male | 38 (76.0) | 30 (66.7) | 68 (54.8) a | |
| Age, years | 75 (66–84) | 84 (75–89) b | 71 (58–83) | |
| Smoking, n (%) | 19 (38.0) | 16 (35.6) | 6 (4.8) b,d | |
| Alcohol intake, n (%) | 28 (56.0) | 7 (15.5) b | 6 (4.8) b,c | |
| Comorbidities | ||||
| Type 2 diabetes mellitus, n (%) | 0 | 22 (48.9) | 30 (23.8) e | |
| Cardiovascular disease, n (%) | 0 | 18 (40) | 68 (54) | |
| Chronic liver disease, n (%) | 0 | 0 | 1 (0.8) | |
| Chronic lung disease, n (%) | 0 | 0 | 18 (14.3) | |
| Chronic kidney disease, n (%) | 0 | 19 (42.2) | 22 (17.5) d | |
| Chronic neurological disease n (%), | 0 | 0 | 29 (23) | |
| Cancer, n (%) | 0 | 17 (37.8) | 16 (12.7) e | |
| Charlson Index | No comorbidity, n (%) | NA | 10 (22.2) | 83 (65.9) e,* |
| Low comorbidity, n (%) | 18 (40.0) | 29 (23.0) | ||
| High comorbidity, n (%) | 17 (37.8) | 14 (11.1) | ||
| McCabe Index | RFD, n (%) | NA | 10 (22.2) | 7 (5.6) e, * |
| UFD, n (%) | 19 (42.2) | 31 (24.6) | ||
| NFD, n (%) | 16 (35.6) | 88 (69.8) | ||
| Medications | ||||
| ACEIs, n (%) | NA | 14 (31.1) | 24 (27.0) | |
| ARAs, n (%) | NA | 12 (26.7) | 21 (16.7) | |
| Oral antidiabetics, n (%) | NA | 19 (42.2) | 37 (29.4) | |
| Insulin, n (%) | NA | 9 (20.0) | 28 (22.2) | |
| Statins, n (%) | NA | 16 (35.6) | 44 (34.9) | |
| Variable | Number of Cases (%) |
|---|---|
| Admission to Intensive Care Unit | 22 (17.5) |
| Non-invasive mechanical ventilation | 6 (4.8) |
| Invasive mechanical ventilation | 20 (15.9) |
| High-flow oxygen therapy | 10 (7.9) |
| Conventional oxygen therapy | 92 (73.0) |
| Deceased | 29 (23.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Tomàs, E.; Iftimie, S.; Castañé, H.; Baiges-Gaya, G.; Hernández-Aguilera, A.; González-Viñas, M.; Castro, A.; Camps, J.; Joven, J. Clinical Performance of Paraoxonase-1-Related Variables and Novel Markers of Inflammation in Coronavirus Disease-19. A Machine Learning Approach. Antioxidants 2021, 10, 991. https://doi.org/10.3390/antiox10060991
Rodríguez-Tomàs E, Iftimie S, Castañé H, Baiges-Gaya G, Hernández-Aguilera A, González-Viñas M, Castro A, Camps J, Joven J. Clinical Performance of Paraoxonase-1-Related Variables and Novel Markers of Inflammation in Coronavirus Disease-19. A Machine Learning Approach. Antioxidants. 2021; 10(6):991. https://doi.org/10.3390/antiox10060991
Chicago/Turabian StyleRodríguez-Tomàs, Elisabet, Simona Iftimie, Helena Castañé, Gerard Baiges-Gaya, Anna Hernández-Aguilera, María González-Viñas, Antoni Castro, Jordi Camps, and Jorge Joven. 2021. "Clinical Performance of Paraoxonase-1-Related Variables and Novel Markers of Inflammation in Coronavirus Disease-19. A Machine Learning Approach" Antioxidants 10, no. 6: 991. https://doi.org/10.3390/antiox10060991