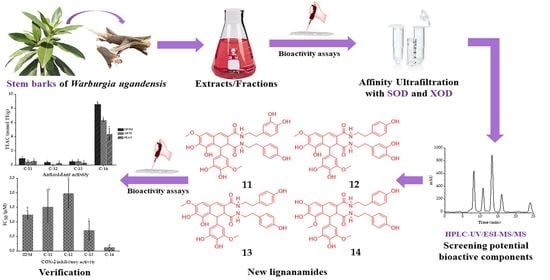
New Lignanamides with Antioxidant and Anti-Inflammatory Activities Screened Out and Identified from Warburgia ugandensis Combining Affinity Ultrafiltration LC-MS with SOD and XOD Enzymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Chemicals and Reagents
2.3. Plant Materials
2.4. Preparation of Samples
2.4.1. Preparation of Extractions and Fractions
2.4.2. Isolation of Pure Compounds
2.5. In Vitro Antioxidant Assays of Samples
2.5.1. DPPH Assays
2.5.2. ABTS Assays
2.5.3. FRAP Assays
2.6. In Vitro COX-2 Inhibitory Assays
2.7. Screening and Identification of the Potential Ligands of SOD and XOD with UF-LC-MS/MS
2.7.1. Affinity Ultrafiltration with SOD and XOD
2.7.2. HPLC-UV/ESI-MS/MS Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Antioxidant Activities of Different Extracts of Stem Barks from W. ugandensis
3.2. Antioxidant Activities of Different Fractions Eluted from WUE
3.3. Screening and Identification of SOD and XOD Ligands by UF-LC-MS/MS
3.3.1. Screening for the Potential SOD and XOD Ligands in WUE-A4
3.3.2. Compounds Isolated and Identified from WUE-A4
3.4. Antioxidant Activities of Compounds Isolated from WUE-A4
3.5. Anti-inflammatory Activities of Compounds Isolated from WUE-A4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drage, S.; Mitter, B.; Tröls, C.; Muchugi, A.; Jamnadass, R.H.; Sessitsch, A.; Hadacek, F. Antimicrobial drimane sesquiterpenes and their effect on endophyte communities in the medical tree Warburgia ugandensis. Front. Microbiol. 2014, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Leonard, C.M.; Viljoen, A.M. Warburgia: A comprehensive review of the botany, traditional uses and phytochemistry. J. Ethnopharmacol. 2015, 165, 260–285. [Google Scholar] [CrossRef]
- Maroyi, A. The genus Warburgia: A review of its traditional uses and pharmacology. Pharm. Biol. 2014, 52, 378–391. [Google Scholar] [CrossRef] [Green Version]
- Mudyiwa, M.; Rajab, M.S.; Fronczek, F.R.; Watkins, S.F. (1R,4R,5aS,7S,9aS)-7,9a-Dimethyl-6-methyl-ene-3-oxo-1,3,4,5,5a,6,7,8,9,9a-deca-hydro-naphtho-[1,2-c]furan-1,4-diyl diacetate. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, 2612–2613. [Google Scholar] [CrossRef] [Green Version]
- Opiyo, S.A.; Manguro, L.O.A.; Okinda-Owuor, P.; Ateka, E.M.; Lemmen, P. 7α-Acetylugandensolide and antimicrobial properties of Warburgia ugandensis extracts and isolates against sweet potato pathogens. Phytochem. Lett. 2011, 4, 161–165. [Google Scholar] [CrossRef]
- Wube, A.A.; Bucar, F.; Gibbons, S.; Asres, K. Sesquiterpenes from Warburgia ugandensis and their antimycobacterial activity. Phytochemistry 2005, 66, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Litaudon, M.; Krief, S.; Martin, M.T.; Kasenene, J.; Kiremire, B.; Dumontet, V.; Gueritte, F. Ugandenial A, a new drimane-type sesquiterpenoid from Warburgia ugandensis. Molecules 2009, 14, 3844–3850. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Jackson, S.H.; Rajab, M.S.; Fronczek, F.R.; Watkins, S.F. Cinnamolid-3β-ol hemihydrate and 3β-hydroxycinnamolide acetate, two drimanolide-class sesquiterpene lactones from Warburgia ugandensis. Acta Crystallogr. C 2006, 62, 219–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manguro, L.O.A.; Ugi, I.; Lemmen, P.; Hermann, R. Flavonol glycosides of Warburgia ugandensis leaves. Phytochemistry 2003, 64, 891–896. [Google Scholar] [CrossRef]
- Kioy, D.; Gray, A.I.; Waterman, P.G. A comparative study of the stem-bark drimane sesquiterpenes and leaf volatile oils of Warburgia ugandensis and W. Stuhlmannii. Phytochemistry 1990, 29, 3535–3538. [Google Scholar] [CrossRef]
- Mbieda, J.N.; Lissouck, D.; Amoa Onguene, P.P.; Ateba Amana, B.; Moto Ongagna, J.; Toze, F.A.; Bikele Mama, D. Insight into the antioxidant and antiradical properties of colorotane sesquiterpenes extracted from Warburgia ugandensis: Theoretical evaluation. Struct. Chem. 2020. [Google Scholar] [CrossRef]
- Wube, A.A.; Gibbons, S.; Asres, K.; Streit, B.; Adams, M.; Bauer, R.; Bucar, F. In vitro 12(S)-HETE and leukotriene metabolism inhibitory activity of sesquiterpenes of Warburgia ugandensis. Planta Med. 2006, 72, 754–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, I.; Fujita, K.; Lee, S.H.; Ha, T.J. Antibacterial activity of polygodial. Phytother. Res. 2005, 19, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Olila, D.; Opuda-Asibo, J. Screening extracts of Zanthoxylum chalybeum and Warburgia ugandensis for activity against measles virus (Swartz and Edmonston strains) in vitro. Afr. Health Sci. 2001, 1, 66–72. [Google Scholar] [PubMed]
- Irungu, B.N.; Rukunga, G.M.; Mungai, G.M.; Muthaura, C.N. In vitro antiplasmodial and cytotoxicity activities of 14 medicinal plants from Kenya. S. Afr. J. Bot. 2007, 73, 204–207. [Google Scholar] [CrossRef] [Green Version]
- Carocho, M.; Morales, P.; Ferreira, I.C.F.R. Antioxidants: Reviewing the chemistry, food applications, legislation and role as preservatives. Trends Food. Sci. Technol. 2018, 71, 107–120. [Google Scholar] [CrossRef] [Green Version]
- Sugahara, S.; Chiyo, A.; Fukuoka, K.; Ueda, Y.; Tokunaga, Y.; Nishida, Y.; Kinoshita, H.; Matsuda, Y.; Igoshi, K.; Ono, M.; et al. Unique antioxidant effects of herbal leaf tea and stem tea from Moringa oleifera L. especially on superoxide anion radical generation systems. Biosci. Biotechnol. Biochem. 2018, 82, 1973–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martorell, M.; Lucas, X.; Alarcón-Zapata, P.; Capó, X.; Quetglas-Llabrés, M.M.; Tejada, S.; Sureda, A. Targeting xanthine oxidase by natural products as a therapeutic approach for mental disorders. Curr. Pharm. Des. 2020, 26, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, S.; Oh, Y.; Kim, Y.M.; Chin, Y.W.; Cho, J. Inhibition of oxidative neurotoxicity and scopolamine-induced memory impairment by γ-mangostin: In vitro and in vivo evidence. Oxid. Med. Cell. Longev. 2019, 2019, 3640753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taleb, A.; Ahmad, K.A.; Ihsan, A.U.; Qu, J.; Lin, N.; Hezam, K.; Koju, N.; Hui, L.; Qilong, D. Antioxidant effects and mechanism of silymarin in oxidative stress induced cardiovascular diseases. Biomed. Pharmacother. 2018, 102, 689–698. [Google Scholar] [CrossRef]
- Barnes, P.J. Oxidative stress-based therapeutics in COPD. Redox Biol. 2020, 33, 101544. [Google Scholar] [CrossRef] [PubMed]
- Sabahi, Z.; Farmani, F.; Soltani, F.; Moein, M. DNA protection, antioxidant and xanthine oxidase inhibition activities of polyphenol-enriched fraction of Berberis integerrima Bunge fruits. Iran J. Basic Med. Sci. 2018, 21, 411–416. [Google Scholar] [PubMed]
- Knaus, U.G. Oxidants in physiological processes. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–21. [Google Scholar]
- Zhu, M.Z.; Wu, W.; Jiao, L.L.; Yang, P.F.; Guo, M.Q. Analysis of flavonoids in lotus (Nelumbo nucifera) leaves and their antioxidant activity using macroporous resin chromatography coupled with LC-MS/MS and antioxidant biochemical assays. Molecules 2015, 20, 10553–10565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.P.; Chang, S.K.C.; Gu, Y.; Qian, S.Y. Antioxidant activity and phenolic compositions of lentil (Lens culinaris var. Morton) extract and its fractions. J. Agric. Food Chem. 2011, 59, 2268–2276. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.L.; Lee, D.Y.; Shang, Y.; Cao, X.T.; Wang, S.Q.; Liao, J.; Zhang, T.; Dai, R.H. Characterization of spirostanol glycosides and furostanol glycosides from Anemarrhenae rhizoma as dual targeted inhibitors of 5-lipoxygenase and cyclooxygenase-2 by employing a combination of affinity ultrafiltration and HPLC/MS. Phytomedicine 2020, 77, 153284. [Google Scholar] [CrossRef]
- Zhu, H.B.; Liu, S.; Li, X.; Song, F.R.; Liu, Z.Q.; Liu, S.Y. Bioactivity fingerprint analysis of cyclooxygenase-2 ligands from Radix aconiti by ultrafiltration-UPLC-MSn. Anal. Bioanal. Chem. 2013, 405, 7437–7445. [Google Scholar] [CrossRef]
- Chen, G.L.; Xu, Y.B.; Wu, J.L.; Li, N.; Guo, M.Q. Hypoglycemic and hypolipidemic effects of Moringa oleifera leaves and their functional chemical constituents. Food Chem. 2020, 333, 127478. [Google Scholar] [CrossRef]
- Chen, G.L.; Fan, M.X.; Wu, J.L.; Li, N.; Guo, M.Q. Antioxidant and anti-inflammatory properties of flavonoids from lotus plumule. Food Chem. 2019, 277, 706–712. [Google Scholar] [CrossRef]
- Xu, Y.B.; Chen, G.L.; Guo, M.Q. Antioxidant and anti-inflammatory activities of the crude extracts of Moringa oleifera from Kenya and their correlations with flavonoids. Antioxidants 2019, 8, 296. [Google Scholar] [CrossRef] [Green Version]
- Kuglerova, M.; Tesarova, H.; Grade, J.T.; Halamova, K.; Wanyana-Maganyi, O.; Damme, P.V.; Kokoska, L. Antimicrobial and antioxidative effects of Ugandan medicinal barks. Afri. J. Biotechnol. 2011, 10, 3628–3632. [Google Scholar]
- Munigunti, R.; Mulabagal, V.; Calderón, A.I. Screening of natural compounds for ligands to PfTrxR by ultrafiltration and LC-MS based binding assay. J. Pharm. Biomed. Anal. 2011, 55, 265–271. [Google Scholar] [CrossRef]
- Lin, Y.H.; Tsai, J.S.; Chen, G.W. Purification and identification of hypocholesterolemic peptides from freshwater clam hydrolysate with in vitro gastrointestinal digestion. J. Food Biochem. 2017, 41, 12358. [Google Scholar] [CrossRef]
- Meerungrueang, W.; Panichayupakaranant, P. A new antibacterial tetrahydronaphthalene lignanamide, foveolatamide, from the stems of Ficus foveolata. Nat. Prod. Commun. 2016, 11, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Peng, D.; Lin, X.L.; Jiang, L.; Huang, J.; Zeng, G.Y.; Deng, X.; Zhou, Y.J. Five macrocyclic glycosides from Schoenoplectus tabernaemontani. Nat. Prod. Res. 2019, 33, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Chang, G.Y.; Ko, F.N.; Teng, C.M. Bioactive constitutents from the stems of Annona montana. Planta Med. 1995, 61, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, Y.; Harinantenaina, L.; Sugimoto, S.; Matsunami, K.; Otsuka, H. C-Glycosyl flavonoids and coloratane-type sesquiterpene glucosides from the water-soluble fraction of a leaf extract of a Malagasy endemic plant, Cinnamosma fragrans (Canellaceae). J. Nat. Med. 2013, 67, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Do, T.Q.; Truong, B.N.; Mai, H.D.T.; Nguyen, T.L.; Nguyen, V.H.; Nguyen, H.D.; Nguyen, T.D.; Nguyen, T.C.; Luong, T.V.; Giang, L.T.; et al. New dianthramide and cinnamic ester glucosides from the roots of Aconitum carmichaelii. J. Asian Nat. Prod. Res. 2019, 21, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, H.B.; Xu, Z.P.; Cheng, Y.G.; Lv, S.W.; Yang, B.Y.; Guo, H.W.; Kuang, H.X. Chemical constituents of Helicia nilagirica Beed. J. Chin. Pharm. Sci. 2010, 45, 1224–1227. [Google Scholar]
- Medina, R.P.; Araujo, A.R.; Andersen, R.J.; Soares, M.A.; Silva, F.A.; Silva, D.H.S. Aromatic compounds produced by endophytic fungi isolated from red alga Asparagopsis taxiformis—Falkenbergia stage. Nat. Prod. Res. 2019, 33, 443–446. [Google Scholar] [CrossRef] [Green Version]
- Morimura, K.I.; Gatayama, A.; Tsukimata, R.; Matsunami, K.; Otsuka, H.; Hirata, E.; Shinzato, T.; Aramoto, M.; Takeda, Y. 5-O-glucosyldihydroflavones from the leaves of Helicia cochinchinensis. Phytochemistry 2006, 67, 2681–2685. [Google Scholar] [CrossRef]
- Günes, M.; Eryilmaz, R.; Aslan, R.; Taken, K.; Demir, H.; Demir, C. Oxidant-antioxidant levels in patients with bladder tumours. Aging Male 2020. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Murray, M. Using N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) to assay cyclooxygenase activity in vitro. Methods Mol. Biol. 2010, 594, 129–140. [Google Scholar]
- Hong, S.S.; Jeong, W.; Kwon, J.G.; Choi, Y.H.; Ahn, E.K.; Ko, H.J.; Seo, D.W.; Oh, J.S. Phenolic amides from the fruits of Tribulus terrestris and their inhibitory effects on the production of nitric oxide. Bull. Korean Chem. Soc. 2013, 34, 3105–3108. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Lee, J.W.; Jang, H.; Le, T.P.L.; Kim, J.G.; Lee, M.S.; Hong, J.T.; Lee, M.K.; HWang, B.Y. Phenolic amides from Tribulus terrestris and their inhibitory effects on nitric oxide production in RAW 264.7 cells. Arch. Pharmacal Res. 2018, 41, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Xu, W.; Xu, T.Q.; Xia, Z.; Zhang, H.X.; Zhan, Y.P.; Zhou, G.X. Barbaram, a bicyclic neolactam from the root and stem of Lycium barbarum. J. Asian Nat. Prod. Res. 2019, 23, 82–88. [Google Scholar] [CrossRef]







| Sample | DPPH # | ABTS # | FRAP # |
|---|---|---|---|
| IC50 (µg/mL) | IC50 (µg/mL) | mmol Fe2+/g | |
| Trolox | 9.0 ± 0.3 f | 5.9 ± 0.1 e | 16.2 ± 1.3 a |
| WUZ | 18.9 ± 0.3 d | 10.2 ± 0.5 d | 1.5 ± 0.1 c |
| WUP | 219.1 ± 1.4 a | 85.5 ± 8.2 a | 0.3 ± 0.1 f |
| WUE | 17.8 ± 0.3 e | 9.4 ± 0.5 d,e | 5.6 ± 0.3 b |
| WUN | 46.5 ± 0.5 b | 22.5 ± 3.9 c | 1.1 ± 0.1 d |
| WUW | 33.8 ± 0.6 c | 32.8 ± 1.6 b | 0.6 ± 0.1 e |
| Sample | DPPH # | ABTS # | FRAP # |
|---|---|---|---|
| IC50 (µg/mL) | IC50 (µg/mL) | mmol Fe2+/g | |
| WUE-A | 18.4 ± 0.3 g | 10.4 ± 0.5 f | 3.4 ± 0.3 b |
| WUE-B | 16.4 ± 0.3 g | 9.6 ± 0.5 f | 7.6 ± 0.4 a |
| WUE-C | 24.7 ± 0.4 f | 10.0 ± 0.8 f | 2.9 ± 0.7 b |
| WUE-D | 55.6 ± 0.9 d | 19.0 ± 3.0 d | 1.0 ± 0.2 c |
| WUE-E | 35.5 ± 0.5 e | 14.8 ± 1.2 e | 0.6 ± 0.1 cd |
| WUE-F | 62.3 ± 0.8 c | 25.4 ± 0.9 c | 0.6 ± 0.1 c,d |
| WUE-G | 243.5 ± 4.2 a | 82.2 ± 3.8 a | 0.4 ± 0.0 d |
| WUE-H | 139.9 ± 1.4 b | 52.3 ± 4.3 b | 0.4 ± 0.0 c,d |
| No. | LC-MS/MS | UF-RBA # | |||||
|---|---|---|---|---|---|---|---|
| Rt (min) | m/z | MS/MS-Fragments | Compounds | SMILES * | XOD | SOD | |
| 1 | 11.8 | 593 | 593, 446, 428, 393, 369, 353, 338, 326, 310, 297, 284, 230, 187 | 2-[3-[2-O-(6-deoxy-α-L-mannopyranosyl)-β-D-glucopyranosyl]-4,5-dihydroxyphenyl]-5,7-dihydroxy-4H-1-benzopyran-4-one | O=C1C2=C(O)C=C(O)C=C2OC(C3=CC(O)=C(O)C([C@@H]4[C@@H](O[C@@H]5[C@@H](O)[C@@H](O)[C@H](O)[C@@H](C)O5)[C@H](O)[C@@H](O)[C@H](CO)O4)=C3)=C1 | 1.2 ± 0.6 c,d | 1.9 ± 0.1 a,b,c |
| 2 | 14.3 | 609 | 609, 352, 301, 284, 271, 254, 216, 192, 162 | Isomer of 5 | - | 1.8 ± 0.1 b,c | - |
| 3 | 17.5 | 577 | 577, 430, 413, 395, 364, 352, 322, 310, 292, 281, 268, 212, 158, 59 | 2-[3-[2-O-(6-deoxy-α-L-mannopyranosyl)-β-D-glucopyranosyl]-4-hydroxyphenyl]-5,7-dihydroxy-4H-1-benzopyran-4-one | O=C1C2=C(O)C=C(O)C=C2OC(C3=CC=C(O)C([C@@H]4[C@@H](O[C@@H]5[C@@H](O)[C@@H](O)[C@H](O)[C@@H](C)O5)[C@H](O)[C@@H](O)[C@H](CO)O4)=C3)=C1 | 1.0 ± 0.2 d | 2.0 ± 0.1 a,b |
| 4 | 20.6 | 625 | 625, 579, 433, 311, 121, 112 | Unknown | - | 0.6 ± 0.1 d | 1.6 ± 0.3 b,c,d,e |
| 5 | 22.7 | 609 | 609, 563, 500, 461, 391, 361, 328, 298, 137, 108, 90, 62 | 4-[(6′-O-β-D-allopyranosyl)-oxy]-hydroxy-benzoic acid cyclic dimeric inner ester * | O[C@@H]1[C@@H](COC(C2=CC=C(O3)C=C2)=O)O[C@@H](OC4=CC=C(C(OC[C@H]5O[C@@H]3[C@H](O)[C@H](O)[C@@H]5O)=O)C=C4)[C@H](O)[C@@H]1O | 1.2 ± 0.1 c,d | 1.1 ± 0.1 e |
| 6 | 25.0 | 617 | 617, 205, 186, 163, 131, 114, 101 | Unknown | - | 2.1 ± 0.3 b | 1.5 ± 0.4 d,e |
| 7 | 27.5 | 298 | 297, 255, 227, 190, 147, 135, 107 | N-trans-caffeoyltyramine | O=C(NCCC1=CC=C(O)C=C1)/C=C/C2=CC(O)=C(O)C=C2 | 1.1 ± 0.3 c,d | 1.1 ± 0.3 e |
| 8 | 28.5 | 327 | 327, 312, 206, 163, 150, 134 | Unknown | - | 2.4 ± 0.2 b | 1.9 ± 0.5 b,c,d |
| 9 | 29.9 | 671 | 671, 530, 491, 475, 453, 418, 392, 367, 352, 338, 299, 282, 229 | Isomer of 11 and 12 | - | - | - |
| 10 | 30.5 | 471 | 471, 403, 373, 289, 263, 235, 208, 150 | Unknown | - | 2.3 ± 0.4 b | - |
| 11 | 32.1 | 671 | 671, 597, 580, 555, 531, 516, 491, 352, 337, 245, 230, 179 | 1-(3,4-dihydroxy-5-methoxyphenyl)-1,2-dihydroxy-7,8-dihydroxy-N-[(3,4-dihydroxyphenyl)ethyl]-N′-[(4-hydroxyphenyl)ethyl]-6-methoxynaphthalene-2,3-dicarboxamide * | OC1=C(OC)C=C2C(C(C3=CC(O)=C(O)C(OC)=C3)C(C(N([H])CCC4=CC=C(O)C=C4)=O)C(C(N([H])CCC5=CC=C(O)C(O)=C5)=O)=C2)=C1O | - | - |
| 12 | 33.1 | 671 | 671, 588, 531, 516, 490, 368, 352, 337, 260, 231, 178 | 1-(3,4-dihydroxy-5-methoxyphenyl)-1,2-dihydroxy-7,8-dihydroxy-N-[(4-hydroxyphenyl)ethyl]-N′-[(4-hydroxyphenyl)ethyl]-6-methoxynaphthalene-2,3-dicarboxamide * | OC1=C(OC)C=C2C(C(C3=CC(O)=C(O)C(OC)=C3)C(C(N([H])CCC4=CC=C(O)C(O)=C4)=O)C(C(N([H])CCC5=CC=C(O)C=C5)=O)=C2)=C1O | 1.7 ± 0.5 b,c | - |
| 13 | 36.1 | 655 | 655, 514, 491, 477, 392, 364, 336, 312, 175 | 1-(3,4-dihydroxy-5-methoxyphenyl)-1,2-dihydroxy-7,8-dihydroxy-N,N′-bis-[2-(4-hydroxyphenyl)ethyl]-6-methoxynaphthalene-2,3-dicarboxamide * | OC1=C(OC)C=C2C(C(C3=CC(O)=C(O)C(OC)=C3)C(C(N([H])CCC4=CC=C(O)C=C4)=O)C(C(N([H])CCC5=CC=C(O)C=C5)=O)=C2)=C1O | 1.3 ± 0.2 c,d | 1.3 ± 0.1 e |
| 14 | 38.5 | 655 | 655, 514, 500, 476, 440, 402, 392, 363, 351, 336 | 1-(3,4-dihydroxy-5-methoxyphenyl)-1,2-dihydroxy-6,7-dihydroxy-N,N′-bis-[2-(4-hydroxyphenyl)ethyl]-8-methoxynaphthalene-2,3-dicarboxamide * | OC1=C(O)C(OC)=C(C(C2=CC(O)=C(O)C(OC)=C2)C(C(N([H])CCC3=CC=C(O)C=C3)=O)C(C(N([H])CCC4=CC=C(O)C=C4)=O)=C5)C5=C1 | 3.8 ± 0.5 a | 1.5 ± 0.1 c,d,e |
| Compounds | DPPH # | ABTS # | FRAP # |
|---|---|---|---|
| IC50 (µM) | IC50 (µM) | mmol Fe2+/g | |
| Trolox | 36.0 ± 1.1 b | 22.4 ± 0.6 c | 16.2 ± 1.3 a |
| WUE-A4 | 10.5 ± 0.7 b,* | 9.2 ± 1.2 c,* | 7.1 ± 0.6 b |
| 1 | 25.8 ± 1.4 b | 62.5 ± 4.2 c | 5.4 ± 0.7 b |
| 7 | 635.8 ± 289.5 a | 462.7 ± 157.3 b | 1.9 ± 0.1 c |
| 11 | 59.1 ± 4.1 b | 80.0 ± 10.3 c | 2.2 ± 0.1 c |
| 12 | 142.5 ± 36.1 b | 1151.8 ± 629.7 a | 1.1 ± 0.2 c |
| 13 | 114.9 ± 20.8 b | 69.8 ± 6.3 c | 1.3 ± 0.2 c |
| 14 | 6.4 ± 0.4 b | 5.4 ± 0.1c | 17.5 ± 1.6 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, X.-C.; Chen, G.-L.; Liu, Y.; Zhang, Y.-L.; Guo, M.-Q. New Lignanamides with Antioxidant and Anti-Inflammatory Activities Screened Out and Identified from Warburgia ugandensis Combining Affinity Ultrafiltration LC-MS with SOD and XOD Enzymes. Antioxidants 2021, 10, 370. https://doi.org/10.3390/antiox10030370
Zhuang X-C, Chen G-L, Liu Y, Zhang Y-L, Guo M-Q. New Lignanamides with Antioxidant and Anti-Inflammatory Activities Screened Out and Identified from Warburgia ugandensis Combining Affinity Ultrafiltration LC-MS with SOD and XOD Enzymes. Antioxidants. 2021; 10(3):370. https://doi.org/10.3390/antiox10030370
Chicago/Turabian StyleZhuang, Xiao-Cui, Gui-Lin Chen, Ye Liu, Yong-Li Zhang, and Ming-Quan Guo. 2021. "New Lignanamides with Antioxidant and Anti-Inflammatory Activities Screened Out and Identified from Warburgia ugandensis Combining Affinity Ultrafiltration LC-MS with SOD and XOD Enzymes" Antioxidants 10, no. 3: 370. https://doi.org/10.3390/antiox10030370