Sorghum Growth Promotion by Paraburkholderia tropica and Herbaspirillum frisingense: Putative Mechanisms Revealed by Genomics and Metagenomics
Abstract
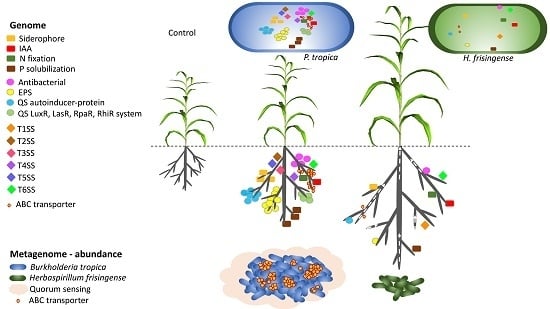
:1. Introduction
2. Materials and Methods
2.1. Bacterial Genome Sequencing and Assembly
2.2. Experimental Design
2.3. Plant Growth Conditions
2.4. Bacterial Isolation, Growth, and Inoculation
2.5. Root Architecture and Plant Biomass Assessment
2.6. DNA Extraction and Shotgun Sequencing
2.7. Shotgun Metagenomic Analysis and Quality Control
2.8. Taxonomic and Functional Analyses
2.9. Normalization
2.10. Potential Bacterial Interactions
2.11. PGPR Traits in the P. Tropica and H. frisingense Genomes
PGPR Gene Homologues, EPS Gene Clusters, Phytohormone Production, and ABC Transporters
3. Results
3.1. Bacterial Genome Assembly and Annotation
3.2. Plant Biomass and Root Architecture
3.3. Metagenomic Sequencing and Read Processing
3.4. Taxonomic Analysis of the Metagenomes
3.5. Functional Analysis of the Metagenomes
3.6. PGPR Gene Homologues
3.7. EPS Gene Clusters
3.8. Phytohormone Production
3.9. PGPR Pathway Analysis
3.10. Community Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marschner, P.; Crowley, D.; Yang, C.H. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 2004, 261, 199–208. [Google Scholar] [CrossRef]
- Fang, M.; Kremer, R.J.; Motavalli, P.P.; Davis, G. Bacterial diversity in rhizospheres of nontransgenic and transgenic corn. Appl. Environ. Microbiol. 2005, 71, 4132–4136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Kuramae, E.E.; de Hollander, M.; Klinkhamer, P.G.; van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017, 11, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, T.R.; Leite, M.F.A.; Lucheta, A.R.; Shimels, M.; Bouwmeester, H.J.; van Veen, J.A.; Kuramae, E.E. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [Green Version]
- van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the Rhizosphere: Tapping into Belowground Chemical Communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef]
- Li, X.; Jousset, A.; de Boer, W.; Carrión, V.J.; Zhang, T.; Wang, X.; Kuramae, E.E. Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 2018, 13, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Dakora, F.D.; Phillips, D.A. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 2002, 245, 35–47. [Google Scholar] [CrossRef]
- Mandimba, G.; Heulin, T.; Bally, R.; Guckert, A.; Balandreau, J. Chemotaxis of free-living nitrogen-fixing bacteria towards maize mucilage. Plant Soil 1986, 90, 129–139. [Google Scholar] [CrossRef]
- Benizri, E.; Baudoin, E.; Guckert, A. Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol Sci. Technol. 2001, 11, 557–574. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial Biofilms: Development, Dispersal, and Therapeutic Strategies in the Dawn of the Postantibiotic Era. Cold Spring Harb. Perspect. Med. 2013, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Sahgal, M.; Johri, B.N. Microbial communication in the rhizosphere: Operation of quorum sensing. Curr. Sci. 2003, 85, 1164–1172. [Google Scholar]
- Seneviratne, G.; Weerasekara, M.; Seneviratne, K.; Zavahir, J.; Kecskés, M.; Kennedy, I. Importance of biofilm formation in plant growth promoting rhizobacterial action. In Plant Growth and Health Promoting Bacteria; Springer: Berlin/Heidelberg, Germany, 2010; pp. 81–95. [Google Scholar]
- Kim, J.; Rees, D.C. Nitrogenase and biological nitrogen fixation. Biochemistry 1994, 33, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.; Siefert, J.L.; Staples, C.R.; Blankenship, R.E. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004, 21, 541–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.S.; Zaidi, A.; Wani, P.A.; Oves, M. Role of plant growth promoting rhizobacteria in the remediation of metal contaminated soils. Environ. Chem. Lett. 2009, 7, 1–19. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Jha, D. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Pagliari, P.H.; Kaiser, D.E.; Rosen, C.J.; Lamb, J.A. The Nature of Phosphorus in Soils; FO-6795-C; University of Minnesota Extension: St Paul, MN, USA, 2017. [Google Scholar]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Holmström, S.J. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar] [PubMed]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef] [PubMed]
- McSteen, P.; Malcomber, S.; Skirpan, A.; Wu, X.; Kellogg, E.; Hake, S. barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 2007, 144, 1000–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010, 12, a001537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patten, C.L.; Glick, B.R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef]
- Arshad, M.; Saleem, M.; Hussain, S. Perspectives of bacterial ACC deaminase in phytoremediation. TRENDS Biotechnol. 2007, 25, 356–362. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mounde, L.G. Understanding the Role of Plant Growth Promoting Bacteria on Sorghum Growth and Biotic Suppression of Striga Infestation. Ph.D. Thesis, University of Hohenheim, Stuttgart, Germany, 2015. [Google Scholar]
- Meena, M.K.; Gupta, S.; Datta, S. Antifungal potential of PGPR, their growth promoting activity on seed germination and seedling growth of winter wheat and genetic variabilities among bacterial isolates. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 235–243. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. Harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef] [Green Version]
- Kirchhof, G.; Eckert, B.; Stoffels, M.; Baldani, J.I.; Reis, V.M.; Hartmann, A. Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int. J. Syst. Evol. Microbiol. 2001, 51, 157–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada, G.A.; Baldani, V.L.D.; de Oliveira, D.M.; Urquiaga, S.; Baldani, J.I. Selection of phosphate-solubilizing diazotrophic Herbaspirillum and Burkholderia strains and their effect on rice crop yield and nutrient uptake. Plant Soil 2013, 369, 115–129. [Google Scholar] [CrossRef]
- Pereira, T.P.; Do Amaral, F.P.; Dall’Asta, P.; Brod, F.C.A.; Arisi, A.C.M. Real-time PCR quantification of the plant growth promoting bacteria Herbaspirillum seropedicae strain SmR1 in maize roots. Mol. Biotechnol. 2014, 56, 660–670. [Google Scholar] [CrossRef] [PubMed]
- da Silva, P.R.A.; Vidal, M.S.; de Paula Soares, C.; Polese, V.; Simões-Araújo, J.L.; Baldani, J.I. Selection and evaluation of reference genes for RT-qPCR expression studies on Burkholderia tropica strain Ppe8, a sugarcane-associated diazotrophic bacterium grown with different carbon sources or sugarcane juice. Antonie Van Leeuwenhoek 2016, 109, 1493–1502. [Google Scholar] [CrossRef]
- da Silveira, A.P.D.; Iório, R.d.P.F.; Marcos, F.C.C.; Fernandes, A.O.; de Souza, S.A.C.D.; Kuramae, E.E.; Cipriano, M.A.P. Exploitation of new endophytic bacteria and their ability to promote sugarcane growth and nitrogen nutrition. Antonie Van Leeuwenhoek 2018. [Google Scholar] [CrossRef]
- Schlemper, T.R.; Dimitrov, M.R.; Silva Gutierrez, F.A.O.; van Veen, J.A.; Silveira, A.P.D.; Kuramae, E.E. Effect of Burkholderia tropica and Herbaspirillum frisingense strains on sorghum growth is plant genotype dependent. PeerJ 2018, 6, e5346. [Google Scholar] [CrossRef] [Green Version]
- Cipriano, M.A.; Lupatini, M.; Lopes-Santos, L.; da Silva, M.J.; Roesch, L.F.; Destéfano, S.A.; Freitas, S.S.; Kuramae, E.E. Lettuce and rhizosphere microbiome responses to growth promoting Pseudomonas species under field conditions. FEMS Microbiol. Ecol. 2016, 92, fiw197. [Google Scholar] [CrossRef] [Green Version]
- Armada, E.; Leite, M.F.A.; Medina, A.; Azcón, R.; Kuramae, E.E. Native bacteria promote plant growth under drought stress condition without impacting the rhizomicrobiome. FEMS Microbiol. Ecol. 2018, 94, fiy092. [Google Scholar] [CrossRef] [Green Version]
- de Boer, W. Upscaling of fungal–bacterial interactions: From the lab to the field. Curr. Opin. Microbiol. 2017, 37, 35–41. [Google Scholar] [CrossRef]
- Velmourougane, K.; Saxena, G.; Prasanna, R. Plant-microbe interactions in the rhizosphere: Mechanisms and their ecological benefits. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; pp. 193–219. [Google Scholar]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotation using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Xing, S.; Ma, H.; Du, Z.; Ma, B. Plant growth-promoting rhizobacteria affect the growth and nutrient uptake of Fraxinus americana container seedlings. Appl. Microbiol. Biotechnol. 2013, 97, 4617–4625. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. In Circular; California Agricultural Experiment Station: Berkeley, CA, USA, 1950; Volume 347. [Google Scholar]
- Mishra, V.; Gupta, A.; Kaur, P.; Singh, S.; Singh, N.; Gehlot, P.; Singh, J. Synergistic effects of arbuscular mycorrhizal fungi and plant growth promoting rhizobacteria in bioremediation of iron contaminated soils. Int. J. Phytoremediation 2016, 18, 697–703. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [Green Version]
- Breitwieser, F.P.; Salzberg, S.L. KrakenHLL: Confident and fast metagenomics classification using unique k-mer counts. BioRxiv 2018, 262956. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- Levy, A.; Gonzalez, I.S.; Mittelviefhaus, M.; Clingenpeel, S.; Paredes, S.H.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138. [Google Scholar] [CrossRef] [Green Version]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2013, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1. 0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plewniak, F.; Koechler, S.; Navet, B.; Dugat-Bony, É.; Bouchez, O.; Peyret, P.; Seby, F.; Battaglia-Brunet, F.; Bertin, P.N. Metagenomic insights into microbial metabolism affecting arsenic dispersion in Mediterranean marine sediments. Mol. Ecol. 2013, 22, 4870–4883. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Pollard, K.S. Average genome size estimation improves comparative metagenomics and sheds light on the functional ecology of the human microbiome. Genome Biol. 2015, 16, 51. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Huang, H.; Wu, C.H. Protein bioinformatics databases and resources. In Protein Bioinformatics; Humana Press: New York, NY, USA, 2017; pp. 3–39. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2013, 42, D199–D205. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Bernabeu, P.R.; Pistorio, M.; Torres-Tejerizo, G.; Estrada-De los Santos, P.; Galar, M.L.; Boiardi, J.L.; Luna, M.F. Colonization and plant growth-promotion of tomato by Burkholderia tropica. Sci. Hortic. 2015, 191, 113–120. [Google Scholar] [CrossRef]
- Zarate, A.; Florez, J.; Angulo, E.; Varela-Prieto, L.; Infante, C.; Barrios, F.; Barraza, B.; Gallardo, D.; Valdes, J. Burkholderia tropica as a potential microalgal growth-promoting bacterium in the biosorption of mercury from aqueous solutions. J. Microbiol. Biotechnol. 2017, 27, 1138–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straub, D.; Yang, H.; Liu, Y.; Tsap, T.; Ludewig, U. Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30T. J. Exp. Bot. 2013, 64, 4603–4615. [Google Scholar] [CrossRef] [Green Version]
- Straub, D.; Rothballer, M.; Hartmann, A.; Ludewig, U. The genome of the endophytic bacterium H. frisingense GSF30T identifies diverse strategies in the Herbaspirillum genus to interact with plants. Front. Microbiol. 2013, 4, 168. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Ibanez, S.; Chini, A.; Solano, R. How microbes twist jasmonate signaling around their little fingers. Plants 2016, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.; Yeoh, Y.K.; Donose, B.C.; Webb, R.I.; Parsons, J.; Liao, W.; Sagulenko, E.; Lakshmanan, P.; Hugenholtz, P. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 2016, 6, 37389. [Google Scholar] [CrossRef]







| P. tropica IAC/BECa 135 | H. frisingense IAC/BECa 152 | |
|---|---|---|
| Total length (Mb) | 8.82 | 5.55 |
| Number of chromosomes (chromids) | 1 (4) | 1 |
| Number of predicted genes | 7711 | 5009 |
| Number of annotated genes | 6929 | 4682 |
| Fraction annotated | 0.89 | 0.93 |
| Pt-1 | Pt-2 | Pt-2 | Hf-1 | Hf-2 | Hf-3 | C-1 | C-2 | C-3 | |
|---|---|---|---|---|---|---|---|---|---|
| Assembled contigs | 222,198 | 133,137 | 208,350 | 288,324 | 258,768 | 291,941 | 323,818 | 303,981 | 307,497 |
| Average length (bp) | 618 | 633 | 608 | 692 | 650 | 661 | 660 | 645 | 639 |
| Total length (Mb) | 137.3 | 84.2 | 126.6 | 199.5 | 168.2 | 192.8 | 213.9 | 196.0 | 196.6 |
| Predicted genes | 565,399 | 400,139 | 539,756 | 825,928 | 719,776 | 832,996 | 849,382 | 806,712 | 822,576 |
| Fraction complete | 0.06 | 0.06 | 0.06 | 0.07 | 0.06 | 0.06 | 0.07 | 0.07 | 0.06 |
| Annotated genes | 244,718 | 194,797 | 232,699 | 372,477 | 323,061 | 378,455 | 387,321 | 347,391 | 352,764 |
| Fraction annotated | 0.43 | 0.49 | 0.43 | 0.45 | 0.45 | 0.45 | 0.46 | 0.43 | 0.43 |
| Cluster | Export Protein | CAZY Families Present | Homologous Clusters |
|---|---|---|---|
| Pt-C-1 | MATE efflux protein, outer membrane polysaccharide export protein | GH39, GT4, GT2 | Colanic acid |
| Pt-C-2 | ABC transporter protein | GT2, GT4 | None |
| Pt-C-3 | ABC transporter protein | GT21, GT5 | None |
| Pt-C-4 | MATE efflux protein | None found | Cepacian |
| Pt-C-5 | RND efflux protein | GT1, GT2, CE14, GT2 | None |
| Pt-C-6 | Polysaccharide export protein | GT2, GT4 | Cepacian |
| Pt-C-7 | Polysaccharide export protein | GT2, GT4, GH5 | O-antigen |
| Hf-C-1 | Polysaccharide export protein | GT2, GT4, GT9, GT11, GT28, GT30, GH109 | O-antigen |
| Hf-C-2 | Polysaccharide export protein | GT2, GT4, GT26 | None |
| Monosaccharide Transporters | IAC/BECa 135 | IAC/BECa 152 |
|---|---|---|
| Glucose/Arabinose | - | - |
| Glucose/Manose | + | + |
| Ribose/D-xylose | + | + |
| L-arabinose | + | - |
| Methyl-galactoside | - | - |
| D-xylose | + | + |
| D-allose | - | - |
| Fructose | + | - |
| Rhamnose | + | - |
| Erythritol | + | - |
| Xylitol | + | - |
| Myo-inositol | + | + |
| Myo-inositol 1-phosphate | - | - |
| Glycerol | - | + |
| Sn-glycerol 3-phosphate | - | + |
| Total | 9 | 6 |
| Phosphate and amino acid transporters | ||
| Phosphate | + | + |
| Phosphonate | + | + |
| Lysine/Arginine/Omithine | + | - |
| Histidine | + | - |
| Glutamine | - | - |
| Arginine | - | - |
| Glutamate/Aspartate | + | + |
| Octopine/Nopaline | + | - |
| General L-amino acid | + | + |
| Glutamate | - | - |
| Cystine | + | + |
| Arginine/Omithine | + | - |
| Lysine | - | - |
| Branched amino acid | + | + |
| Neutral amino acid | - | - |
| Urea | + | + |
| D-methionine | - | - |
| Total | 11 | 7 |
| Other transporters | ||
| Oligopeptide | + | - |
| Gluthathione | - | + |
| Iron complex | + | + |
| Lipopolysaccharide | + | - |
| Lipo-oligosaccharide | + | - |
| Total | 4 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuramae, E.E.; Derksen, S.; Schlemper, T.R.; Dimitrov, M.R.; Costa, O.Y.A.; da Silveira, A.P.D. Sorghum Growth Promotion by Paraburkholderia tropica and Herbaspirillum frisingense: Putative Mechanisms Revealed by Genomics and Metagenomics. Microorganisms 2020, 8, 725. https://doi.org/10.3390/microorganisms8050725
Kuramae EE, Derksen S, Schlemper TR, Dimitrov MR, Costa OYA, da Silveira APD. Sorghum Growth Promotion by Paraburkholderia tropica and Herbaspirillum frisingense: Putative Mechanisms Revealed by Genomics and Metagenomics. Microorganisms. 2020; 8(5):725. https://doi.org/10.3390/microorganisms8050725
Chicago/Turabian StyleKuramae, Eiko E., Stan Derksen, Thiago R. Schlemper, Maurício R. Dimitrov, Ohana Y. A. Costa, and Adriana P. D. da Silveira. 2020. "Sorghum Growth Promotion by Paraburkholderia tropica and Herbaspirillum frisingense: Putative Mechanisms Revealed by Genomics and Metagenomics" Microorganisms 8, no. 5: 725. https://doi.org/10.3390/microorganisms8050725