GroEL Secreted from Bacillus subtilis Natto Exerted a Crucial Role for Anti-Inflammatory IL-10 Induction in THP-1 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Fermentation
2.2. Cytokine Measurement
2.3. Purification of the Immunomodulatory Component
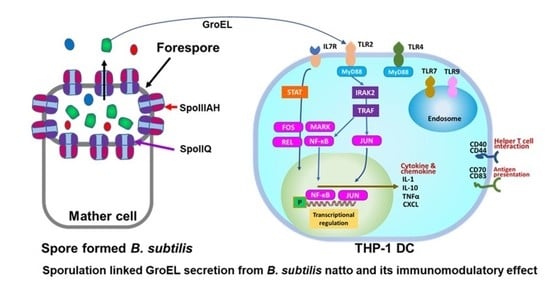
2.4. Sporulation of B. subtilis Natto Strains
2.5. SDS-PAGE, Western Blotting, and Protein Identification
2.6. Gene Expression Analysis for B. subtilis Natto 1 and 15
2.7. Gene Expression Analysis for THP-1 DC Treated with GroEL
2.8. Statistical Analysis
3. Results
3.1. Isolation of Bacillus subtilis Natto from Natto Products
3.2. Cytokine Inducing Activity
3.3. Purification of the Active Components
3.4. Western Blotting Analysis and Inhibition of IL-10-Inducing Activity
3.5. Prediction of TLRs for GroEL Interaction
3.6. Differentially Expressed Genes in THP-1 DCs after GroEL Treatment
3.7. Induced Gene Expressions in B. subtilis natto 1
3.8. GroEL Secretion in Spored Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pärtty, A.; Rautava, S.; Kalliomäki, M. Probiotics on pediatric functional gastrointesti nal disorders. Nutrients 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.; Sartor, R.B. Probiotics and inflammatory bowel diseases. Am. J. Gastroenterol. 2000, 95, S19–S21. [Google Scholar] [CrossRef]
- Salminen, S.; Isolauri, E.; Onnela, T. Gut flora in normal and disordered states. Chemotherapy 1995, 41, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Macfarlan, G.T.; Cummings, J.H. Probiotics, infection and immunity, Curr. Opin. Infect. Dis. 2002, 15, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; van Hylckama Vlieg, J.E. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 2011, 10, S1–S3. [Google Scholar] [CrossRef]
- Llewellyn, A.; Foey, A. Probiotic modulation of innate cell pathogen sensing and signaling events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Huang, A.Y.; Germain, R.N. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med. 2006, 203, 2841–2852. [Google Scholar]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Thomas, C.M.; Versalovic, J. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbe. 2010, 1, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, F.A.; Yamamoto, H.; Kobayashi, Y.; Sekiguchi, J. Characterization of a new sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 1999, 181, 6230–6237. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Tani, K.; Soda, K.; Misono, H. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-gamma-glutamate. J. Biochem. 1998, 123, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Duc, L.H.; Cutting, S.M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Hosoi, T.; Kiuchi, K. Production and probiotic effects of natto. In Horizon Bioscience; Ricca, E., Henriques, A.O., Cutting, S.M., Eds.; Horizon Bioscience: Wymondham, UK, 2004. [Google Scholar]
- Hosoi, T.; Ametani, A.; Kiuchi, K.; Kaminogawa, S. Changes in fecal microflora induced by intubation of mice with Bacillus subtilis (natto) spores are dependent upon dietary components. Can. J. Microbiol. 1999, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hosoi, T.; Ametani, A.; Kiuchi, K.; Kaminogawa, S. Improved growth and viability of lactobacilli in the presence of Bacillus subtilis (natto), catalase, or subtilisin. Can. J. Microbiol. 2000, 46, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Inooka, S.; Uehara, S.; Kimura, M. The effect of Bacillus natto on the T and B lymphocytes from spleens of feeding chickens. Poult. Sci. 1986, 65, 1217–1219. [Google Scholar] [CrossRef] [PubMed]
- Samanya, M.; Yamauchi, K.E. Histological alterations of intestinal villi in chickens fed dried Bacillus subtilis var. natto. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 133, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Cavaillon, J.M. Cytokines and macrophages. Biomed. Pharmacother. 1994, 48, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.T.; Chen, T.L.; Chen, R.M. Lipopolysaccharide triggers macrophage activation of inflammatory cytokine expression, chemotaxis, phagocytosis, and oxidative ability via a toll-likereceptor 4-dependent pathway: Validated by RNA interference. Toxicol. Lett. 2009, 191, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Cho, S.B.; Lee, H.J.; Chung, W.T.; Kim, K.H.; Hwangbo, J.; Nam, I.S.; Cho, Y.; Yang, M.P.; Chung, I.B. Comparison of cytokine and nitric oxide induction in murine macrophages between whole cell and enzymatically digested Bifidobacterium sp obtained from monogastric animals. J. Microbiol. 2007, 45, 305–310. [Google Scholar] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Lemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- García-Carnero, L.C.; Salinas-Marín, R.; Lozoya-Pérez, N.E.; Wrobel, K.; Wrobel, K.; Martínez-Duncker, I.; Niño-Vega, G.A.; Mora-Montes, H.M. The heat shock protein 60 and Pap1 participate in the Sporothrixschenckii-Host Interaction. J. Fungi 2021, 12, 960. [Google Scholar] [CrossRef] [PubMed]
- Re, F.; Strominger, J.L. Heterogeneity of TLR-induced responses in dendriticcells: From innate to adaptive immunity. Immunobiology 2004, 209, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Guez, J.S.; Vassaux, A.; Larroche, C.; Jacques, P.; Coutte, F. New continuous process for the production of lipopeptide biosurfactants in foam overflowing bioreactor. Front. Bioeng. Biotechnol. 2021, 9, 678469. [Google Scholar] [CrossRef]
- Yin, T.; Zhang, X.; Iwatani, S.; Miyanaga, K.; Yamamoto, N. Uptake of Levilactobacillus brevis JCM 1059 by THP-1 cells via interaction between SlpB and CAP-1 promotes cytokine production. Microorganisms 2023, 11, 247. [Google Scholar] [CrossRef]
- Park, J.S.; Choi, J.W.; Jhun, J.; Kwon, J.Y.; Lee, B.I.; Yang, C.W.; Park, S.H.; Cho, M.L. Lactobacillus acidophilus improves intestinal inflammation in an acute colitis mouse model by regulation of Th17 and Treg cell balance and fibrosis development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef]
- Gopikrishna, T.; Suresh Kumar, H.K.; Perumal, K.; Elangovan, E. Impact of Bacillus in fermented soybean foods on human health. Ann. Microbiol. 2021, 71, 30. [Google Scholar] [CrossRef]
- Tsai, Y.T.; Cheng, P.C.; Pan, T.M. The immunomodulatory effects of lactic acid bacteria for improving immune functions and benefits. Appl. Microbiol. Biotechnol. 2012, 96, 853–862. [Google Scholar] [CrossRef]
- Sung, W.W.; Lin, Y.Y.; Huang, S.D.; Cheng, H.L. Exopolysaccharides of Bacillus amyloliquefaciens Amy-1 mitigate inflammation by inhibiting ERK1/2 and NF-κB pathways and activating p38/Nrf2 pathway. Int. J. Mol. Sci. 2022, 23, 10237. [Google Scholar] [CrossRef]
- Dias, A.M.M.; Douhard, R.; Hermetet, F.; Regimbeau, M.; Lopez, T.E.; Gonzalez, D.; Masson, S.; Marcion, G.; Chaumonnot, K.; Uyanik, B.; et al. Lactobacillus stress protein GroEL prevents colonic inflammation. J. Gastroenterol. 2021, 56, 442–455. [Google Scholar] [CrossRef]
- Khan, M.N.; Shukla, D.; Bansal, A.; Mustoori, S.; Ilavazhagan, G. Immunogenicity and protective efficacy of GroEL (hsp60) of Streptococcus pneumoniae against lethal infection in mice FEMS Immunol. Med. Microbiol. 2009, 56, 56–62. [Google Scholar]
- Craig, E.A.; Gambill, B.D.; Nelson, R.J. Heat shock proteins: Molecular chaperones of protein biogenesis. Microbiol. Rev. 1993, 57, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Shiota, S.; Yamaguchi, N.; Masuhiro, Y.; Hanazawa, S. Induction of Porphyromonas gingivalis GroEL signaling via binding to Toll-like receptors 2 and 4. Oral Microbiol. Immunol. 2006, 21, 245–451. [Google Scholar]
- Camp, A.H.; Losick, R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008, 69, 402–417. [Google Scholar] [CrossRef]
- Doan, T.; Morlot, C.; Meisner, J.; Serrano, M.; Henriques, A.O.; Moran, C.P., Jr.; Rudner, D.Z. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009, 5, e1000566. [Google Scholar] [CrossRef]
- Meisnera, J.; Maehigashib, T.; Andréc, I.; Dunhamb, C.M.; Moran, C.P., Jr. Structure of the basal components of a bacterial transporter. Proc. Natl. Acad. Sci. USA 2012, 109, 5446–5451. [Google Scholar] [CrossRef]
- Katagiri, R.; Sawada, N.; Goto, A.; Yamaji, T.; Iwasaki, M.; Noda, M.; Iso, H.; Tsugane, S.; Japan Public Health Center-based Prospective Study Group. Association of soy and fermented soy product intake with total and cause specific mortality: Prospective cohort study. BMJ 2020, 29, m34. [Google Scholar] [CrossRef]
- Nozue, M.; Shimazu, T.; Charvat, H.; Mori, N.; Mutoh, M.; Sawada, N.; Iwasaki, M.; Yamaji, T.; Inoue, M.; Kokubo, Y.; et al. Fermented soy products intake and risk of cardiovascular disease and total cancer incidence: The Japan Public Health Center-based Prospective study. Eur. J. Clin. Nutr. 2021, 75, 954–968. [Google Scholar] [CrossRef] [PubMed]









| No | Identified Bacterial Strain | Products | Company | Region |
|---|---|---|---|---|
| 1 | Bacillus subtilis (natto) 1 | Kinnotubu natto | Mizkan | Aichi |
| 2 | Bacillus subtilis (natto) 2 | Okame natto | Takanofoods | Ibaraki |
| 3 | Bacillus subtilis (natto) 3 | Kezurikobushi natto | Yamada Foods | Akita |
| 4 | Bacillus subtilis (natto) 4 | Kiriboshi Natto | NUKAGA SYOJI | Ibaraki |
| 5 | Bacillus subtilis (natto) 5 | Natto-jiman | Hoya-Natto | Tokyo |
| 6 | Bacillus subtilis (natto)(Rough) 6 | Sugoi S-903 | Takanofoods | Ibaraki |
| 7 | Bacillus subtilis (natto)(Smooth) 7 | |||
| 8 | Bacillus subtilis (natto) 8 | Tezukuri natto | Shika-ya | Kagoshima |
| 9 | Bacillus subtilis (natto) 9 | Hokkaido Kotubu natto | Kajinoya | Kanagawa |
| 10 | Bacillus subtilis (natto) 10 | Yukihomare | Osato | Ibaraki |
| 11 | Bacillus subtilis (natto) 11 | Kawaguchi Natto | Kawaguchi Natto | Miyagi |
| 12 | Bacillus subtilis (natto) 12 | Hamanatto | Suzukishoji | Shizuoka |
| 13 | Bacillus subtilis (natto) 13 | Tukuba-natto | Tukuba-natto | Ibaraki |
| 14 | Bacillus subtilis (natto) 14 | Ohisama natto | Ibaraki | Ibaraki |
| 15 | Bacillus subtilis (natto) 15 | Tengu natto | Ibaraki | Ibaraki |
| 16 | Bacillus subtilis (natto) 16 | Ibaraki hoshi natto | Ibaraki | Ibaraki |
| 17 | Bacillus subtilis (natto) 17 | Namahoshi natto | Ibaraki | Ibaraki |
| 18 | Bacillus subtilis (natto) 18 | Korumame natto | Musouan | Kumamoto |
| 19 | Bacillus subtilis (natto) 19 | Hikiwari natto | Kamakurayama-Noroshokuhinn | Kanagawa |
| 20 | Bacillus subtilis (natto) 20 | Natto-moto | Takahashiyuzo-Kenkyusho | Yamagata |
| 21 | Bacillus subtilis (natto) 21 | Miyagino natto | Miyagino | Miyagi |
| 22 | Bacillus subtilis (natto) 22 | Funmatsu Natto-kin | Naruse Hakkokagaku Kenkyusho | Tokyo |
| 23 | Bacillus subtilis (natto) 23 | Fukuokajiman | Yoshino Shoten | Fukuoka |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uesugi, T.; Mori, S.; Miyanaga, K.; Yamamoto, N. GroEL Secreted from Bacillus subtilis Natto Exerted a Crucial Role for Anti-Inflammatory IL-10 Induction in THP-1 Cells. Microorganisms 2023, 11, 1281. https://doi.org/10.3390/microorganisms11051281
Uesugi T, Mori S, Miyanaga K, Yamamoto N. GroEL Secreted from Bacillus subtilis Natto Exerted a Crucial Role for Anti-Inflammatory IL-10 Induction in THP-1 Cells. Microorganisms. 2023; 11(5):1281. https://doi.org/10.3390/microorganisms11051281
Chicago/Turabian StyleUesugi, Taisuke, Suguru Mori, Kazuhiko Miyanaga, and Naoyuki Yamamoto. 2023. "GroEL Secreted from Bacillus subtilis Natto Exerted a Crucial Role for Anti-Inflammatory IL-10 Induction in THP-1 Cells" Microorganisms 11, no. 5: 1281. https://doi.org/10.3390/microorganisms11051281