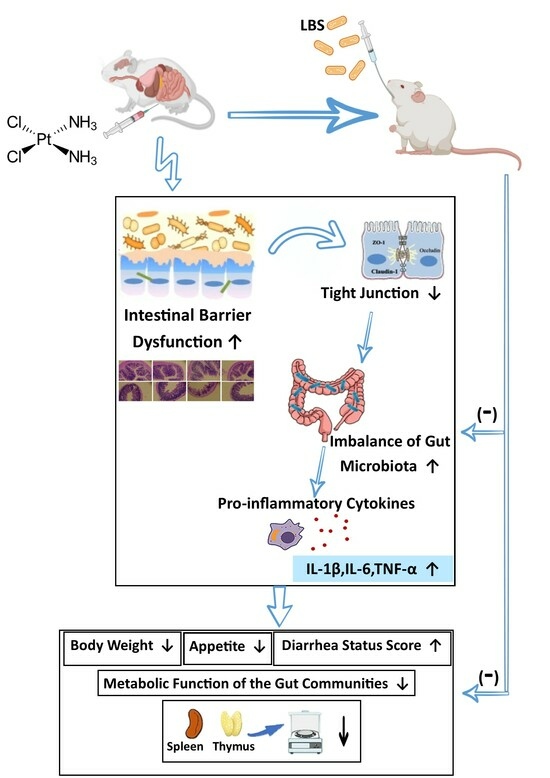
Lactobacillus rhamnosus Attenuates Cisplatin-Induced Intestinal Mucositis in Mice via Modulating the Gut Microbiota and Improving Intestinal Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Probiotic Strain
2.2. Ethical Statement and Experimental Animals
2.3. Study Design
2.4. Measurement of Organ Indices
2.5. Stool Output and Diarrhea Assessment
2.6. Pro-Inflammatory Cytokines Analysis
2.7. Real-Time Quantitative PCR (RT-qPCR)
2.8. Histological Examination
2.9. Mucin Production and Goblet Cells
2.10. Immunofluorescent Staining for Tight Junction Proteins
2.11. Gut Microbiome Genomic DNA Extraction and 16S rRNA Pyrosequencing
2.12. Metagenomic Functional Analysis of the Microbiome Composition
2.13. Statistical Analysis
3. Results
3.1. LBS Treatment Attenuates Body Weight Loss and Increases Food and Water Intake and Organ Index
3.2. LBS Increases Stool Output and Reduces the Severity of Diarrhea
3.3. LBS Attenuates Pro-Inflammatory Cytokine Levels in Cisplatin-Induced Intestinal Mucositis Mice Model
3.4. Effects of LBS on Histopathological Examinations in the Intestinal Mucosal Layer
3.5. LBS Modulates the Tight Junction Protein Expression in the Colon and Ileum of CP-Induced IM Mice
3.6. LBS Treatment Modulates the Gut Microbiota Dysbiosis
3.7. LBS Effect on the Gut Metabolic Functional Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Hajiagha, M.N.; Taghizadeh, S.; Asgharzadeh, M.; Dao, S.; Ganbarov, K.; Köse, Ş.; Kafil, H.S. Gut microbiota and human body interactions; its impact on health: A review. Curr. Pharm. Biotechnol. 2022, 23, 4–14. [Google Scholar] [CrossRef]
- Frost, F.; Weiss, F.U.; Sendler, M.; Kacprowski, T.; Rühlemann, M.; Bang, C.; Franke, A.; Völker, U.; Völzke, H.; Lamprecht, G.; et al. The Gut Microbiome in Patients with Chronic Pancreatitis Is Characterized by Significant Dysbiosis and Overgrowth by Opportunistic Pathogens. Clin. Transl. Gastroenterol. 2020, 11, e00232. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, A.; Zorzi, F.; Del Chierico, F.; Altomare, A.; Cocca, S.; Avola, A.; De Biasio, F.; Russo, A.; Cella, E.; Reddel, S.; et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front. Microbiol. 2019, 10, 1655. [Google Scholar] [CrossRef]
- Madan, A.; Thompson, D.; Fowler, J.C.; Ajami, N.; Salas, R.; Frueh, B.; Bradshaw, M.; Weinstein, B.; Oldham, J.; Petrosino, J. The gut microbiota is associated with psychiatric symptom severity and treatment outcome among individuals with serious mental illness. J. Affect. Disord. 2020, 264, 98–106. [Google Scholar] [CrossRef]
- Geng, J.; Ni, Q.; Sun, W.; Li, L.; Feng, X. The links between gut microbiota and obesity and obesity related diseases. Biomed. Pharmacother. 2022, 147, 112678. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; De La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Sougiannis, A.T.; VanderVeen, B.N.; Davis, J.M.; Fan, D.; Murphy, E.A. Understanding chemotherapy-induced intestinal mucositis and strategies to improve gut resilience. Am. J. Physiol. Liver Physiol. 2021, 320, G712–G719. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef]
- Basile, D.; Di Nardo, P.; Corvaja, C.; Garattini, S.K.; Pelizzari, G.; Lisanti, C.; Bortot, L.; Da Ros, L.; Bartoletti, M.; Borghi, M.; et al. Mucosal Injury during Anti-Cancer Treatment: From Pathobiology to Bedside. Cancers 2019, 11, 857. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Khan, A.Q.; Qamar, W.; Lateef, A.; Tahir, M.; Rehman, M.U.; Ali, F.; Sultana, S. Chrysin protects against cisplatin-induced colon. toxicity via amelioration of oxidative stress and apoptosis: Probable role of p38MAPK and p53. Toxicol. Appl. Pharmacol. 2012, 258, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Freyer, D.R.; Chen, L.; Krailo, M.D.; Knight, K.; Villaluna, D.; Bliss, B.; Pollock, B.H.; Ramdas, J.; Lange, B.; Van Hoff, D.; et al. Effects of sodium thiosulfate versus observation on development of cisplatin-induced hearing loss in children with cancer (ACCL0431): A multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2017, 18, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Livingston, M.J.; Safirstein, R.; Dong, Z. Cisplatin nephrotoxicity: New insights and therapeutic implications. Nat. Rev. Nephrol. 2022, 19, 53–72. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Dasari, S.; Noubissi, F.K.; Ray, P.; Kumar, S. Advances in Our Understanding of the Molecular Mechanisms of Action of Cisplatin in Cancer Therapy. J. Exp. Pharmacol. 2021, 13, 303–328. [Google Scholar] [CrossRef]
- Dugbartey, G.J.; Peppone, L.J.; de Graaf, I.A. An integrative view of cisplatin-induced renal and cardiac toxicities: Molecular mechanisms, current treatment challenges and potential protective measures. Toxicology 2016, 371, 58–66. [Google Scholar] [CrossRef]
- Dahiya, D.; Nigam, P.S. Biotherapy Using Probiotics as Therapeutic Agents to Restore the Gut Microbiota to Relieve Gastro-intestinal Tract Inflammation, IBD, IBS and Prevent Induction of Cancer. Int. J. Mol. Sci. 2023, 24, 5748. [Google Scholar] [CrossRef]
- Nakai, H.; Murosaki, S.; Yamamoto, Y.; Furutani, M.; Matsuoka, R.; Hirose, Y. Safety and efficacy of using heat-killed Lactobacillus plantarum L-137: High-dose and long-term use effects on immune-related safety and intestinal bacterial flora. J. Immunotoxicol. 2021, 18, 127–135. [Google Scholar] [CrossRef]
- Segers, M.E.; Lebeer, S. Towards a better understanding of Lactobacillus rhamnosus GG-host interactions. Microb. Cell Factories 2014, 13 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, Y.; Wan, Y.; Hu, T.; Liu, L.; Yang, S.; Gong, Z.; Zeng, Q.; Wei, Y.; Yang, W.; et al. A Novel Postbiotic From Lactobacillus rhamnosus GG With a Beneficial Effect on Intestinal Barrier Function. Front. Microbiol. 2019, 10, 477. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Ma, T.; Zhang, T.; Jin, H.; Li, Y.; Kwok, L.-Y.; Zhang, H.; Sun, Z. Adjunctive Probiotic Lactobacillus rhamnosus Probio-M9 Administration Enhances the Effect of Anti-PD-1 Antitumor Therapy via Restoring Antibiotic-Disrupted Gut Microbiota. Front. Immunol. 2021, 12, 772532. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Polk, D.B. Probiotics and Probiotic-Derived Functional Factors—Mechanistic Insights into Applications for Intestinal Homeostasis. Front. Immunol. 2020, 11, 1428. [Google Scholar] [CrossRef]
- Shi, Y.; Luo, J.; Narbad, A.; Chen, Q. Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis. Microorganisms 2023, 11, 179. [Google Scholar] [CrossRef]
- Guo, H.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. The Potential Therapeutic Role of Lactobacillaceae rhamnosus for Treatment of Inflammatory Bowel Disease. Foods 2023, 12, 692. [Google Scholar] [CrossRef]
- Chen, L.; Li, S.; Peng, C.; Gui, Q.; Li, J.; Xu, Z.; Yang, Y. Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration. Nutrients 2023, 15, 672. [Google Scholar] [CrossRef]
- Darbandi, A.; Mirshekar, M.; Shariati, A.; Moghadam, M.T.; Lohrasbi, V.; Asadolahi, P.; Talebi, M. The effects of probiotics on reducing the colorectal cancer surgery complications: A periodic review during 2007–2017. Clin. Nutr. 2019, 39, 2358–2367. [Google Scholar] [CrossRef]
- Scalabrin, D.; Harris, C.; Johnston, W.; Berseth, C. Long-term safety assessment in children who received hydrolyzed protein formulas with Lactobacillus rhamnosus GG: A 5-year follow-up. Eur. J. Pediatr. 2016, 176, 217–224. [Google Scholar] [CrossRef]
- Chang, C.-W.; Liu, C.-Y.; Lee, H.-C.; Huang, Y.-H.; Li, L.-H.; Chiau, J.-S.C.; Wang, T.-E.; Chu, C.-H.; Shih, S.-C.; Tsai, T.-H.; et al. Lactobacillus casei Variety rhamnosus Probiotic Preventively Attenuates 5-Fluorouracil/Oxaliplatin-Induced Intestinal Injury in a Syngeneic Colorectal Cancer Model. Front. Microbiol. 2018, 9, 983. [Google Scholar] [CrossRef]
- Barroso FA, L.; de Jesus LC, L.; de Castro, C.P.; Batista, V.L.; Ferreira, Ê.; Fernandes, R.S. Intake of Lactobacillus delbrueckii (pExu:hsp65) Prevents the Inflammation and the Disorganization of the Intestinal Mucosa in a Mouse Model of Mucositis. Microorganisms 2021, 9, 107. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, J.; Lin, Z.; Wang, Q.; Li, Y.; Wang, A.; Shan, X.; Liu, J. Administration of a Probiotic Mixture Ameliorates Cisplatin-Induced Mucositis and Pica by Regulating 5-HT in Rats. J. Immunol. Res. 2021, 2021, 9321196. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.M.; Stringer, A.M.; Gibson, R.J.; Yeoh, A.S.J.; Hannam, S.; Keefe, D.M.K. VSL#3 probiotic treatment reduces chemotherapy-induced diarrhoea and weight loss. Cancer Biol. Ther. 2007, 6, 1449–1454. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kheirelseid, E.A.; Chang, K.H.; Newell, J.; Kerin, M.J.; Miller, N. Identification of endogenous control genes for normalisation of real-time quantitative PCR data in colorectal cancer. BMC Mol. Biol. 2010, 11, 12. [Google Scholar] [CrossRef]
- Katiraei, S.; Anvar, Y.; Hoving, L.; Berbée, J.F.P.; van Harmelen, V.; van Dijk, K.W. Evaluation of Full-Length Versus V4-Region 16S rRNA Sequencing for Phylogenetic Analysis of Mouse Intestinal Microbiota after a Dietary Intervention. Curr. Microbiol. 2022, 79, 276. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 2014, 5, 508. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Louca, S.; Jacques, S.M.S.; Pires, A.P.F.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 2016, 1, 0015. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Jesus, E.C.; Lopes, A.; Aguiar, S.; Begnami, M.D.; Rocha, R.M.; Carpinetti, P.A.; Camargo, A.A.; Hoffmann, C.; Freitas, H.C.; et al. Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling. Front. Cell. Infect. Microbiol. 2016, 6, 179. [Google Scholar] [CrossRef] [PubMed]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in Intestinal Mucosal Defense and Inflammation: Learning from Clinical and Experimental Studies. Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Alhadeff, A.L.; Holland, R.A.; Zheng, H.; Rinaman, L.; Grill, H.J.; De Jonghe, B.C. Excitatory Hindbrain–Forebrain Communication Is Required for Cisplatin-Induced Anorexia and Weight Loss. J. Neurosci. 2016, 37, 362–370. [Google Scholar] [CrossRef]
- Chandrashekar, P.M.; Venkatesh, Y.P. Fructans from aged garlic extract produce a delayed immunoadjuvant response to ovalbumin antigen in BALB/c mice. Immunopharmacol. Immunotoxicol. 2011, 34, 174–180. [Google Scholar] [CrossRef]
- Nakamura, S.; Kuda, T.; An, C.; Kanno, T.; Takahashi, H.; Kimura, B. Inhibitory effects of Leuconostoc mesenteroides 1RM3 isolated from narezushi, a fermented fish with rice, on Listeria monocytogenes infection to Caco-2 cells and A/J mice. Anaerobe 2012, 18, 19–24. [Google Scholar] [CrossRef]
- Gao, D.; Liu, Z.; Liu, F.; Chen, L.; Wang, W.; Ma, J.; Xu, C.; Jiang, Z.; Hou, J. Study of the immunoregulatory effect of Lactobacillus rhamnosus 1.0320 in immunosuppressed mice. J. Funct. Foods 2021, 79, 104423. [Google Scholar] [CrossRef]
- Huang, H.-T.; Hu, Y.-F.; Lee, B.-H.; Huang, C.-Y.; Lin, Y.-R.; Huang, S.-N.; Chen, Y.-Y.; Chang, J.-J.; Nan, F.-H. Dietary of Lactobacillus paracasei and Bifidobacterium longum improve nonspecific immune responses, growth performance, and resistance against Vibrio parahaemolyticus in Penaeus vannamei. Fish Shellfish. Immunol. 2022, 128, 307–315. [Google Scholar] [CrossRef]
- Masood, M.I.; Qadir, M.I.; Shirazi, J.H.; Khan, I.U. Beneficial effects of lactic acid bacteria on human beings. Crit. Rev. Microbiol. 2010, 37, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, B.; Jin, D.; Zhan, M.; Lu, J.; Huo, G. Immunomodulatory activity of Lactobacillus plantarum KLDS1.0318 in cyclophosphamide-treated mice. Food Nutr. Res. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Zhao, X.J.; Wang, J.Y. Synergy of Astragalus polysaccharides and probiotics (Lactobacillus and Bacillus cereus) on immunity and intestinal microbiota in chicks. Poult. Sci. 2009, 88, 519–525. [Google Scholar] [CrossRef]
- Rath, E.; Haller, D. Intestinal epithelial cell metabolism at the interface of microbial dysbiosis and tissue injury. Mucosal Immunol. 2022, 15, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cong, L.; Wang, C.; Li, H.; Zhang, C.; Guan, X.; Liu, P.; Xie, Y.; Chen, J.; Sun, J. Immunomodulatory effect of Schisandra polysaccharides in cyclophosphamide-induced immunocompromised mice. Exp. Ther. Med. 2018, 15, 4755–4762. [Google Scholar] [CrossRef] [PubMed]
- Batista, V.L.; da Silva, T.F.; de Jesus, L.C.L.; Coelho-Rocha, N.D.; Barroso, F.A.L.; Tavares, L.M.; Azevedo, V.; Mancha-Agresti, P.; Drumond, M.M. Probiotics, Prebiotics, Synbiotics, and Paraprobiotics as a Therapeutic Alternative for Intestinal Mucositis. Front. Microbiol. 2020, 11, 544490. [Google Scholar] [CrossRef]
- Quintanilha, M.F.; Miranda, V.C.; Souza, R.O.; Gallotti, B.; Cruz, C.; Santos, E.A.; Alvarez-Leite, J.I.; Jesus, L.C.; Azevedo, V.; Trindade, L.M.; et al. Bifidobacterium longum subsp. longum 51A attenuates intestinal injury against irinotecan-induced mucositis in mice. Life Sci. 2021, 289, 120243. [Google Scholar] [CrossRef]
- Tripathi, P.; Alshahrani, S. Mitigation of IL-1β, IL-6, TNF-α, and markers of apoptosis by ursolic acid against cisplatin-induced oxidative stress and nephrotoxicity in rats. Hum. Exp. Toxicol. 2021, 40 (Suppl. 12), S397–S405. [Google Scholar] [CrossRef]
- Gong, S.; Feng, Y.; Zeng, Y.; Zhang, H.; Pan, M.; He, F.; Wu, R.; Chen, J.; Lu, J.; Zhang, S.; et al. Gut microbiota accelerates cisplatin-induced acute liver injury associated with robust inflammation and oxidative stress in mice. J. Transl. Med. 2021, 19, 147. [Google Scholar] [CrossRef]
- Zeng, X.; Jia, H.; Zhang, X.; Wang, X.; Wang, Z.; Gao, Z.; Yuan, Y.; Yue, T. Supplementation of kefir ameliorates azoxymethane/dextran sulfate sodium induced colorectal cancer by modulating the gut microbiota. Food Funct. 2021, 12, 11641–11655. [Google Scholar] [CrossRef]
- Song, W.; Yang, X.; Wang, W.; Wang, Z.; Wu, J.; Huang, F. Sinomenine ameliorates septic acute lung injury in mice by modulating gut homeostasis via aryl hydrocarbon receptor/Nrf2 pathway. Eur. J. Pharmacol. 2021, 912, 174581. [Google Scholar] [CrossRef]
- Slifer, Z.M.; Blikslager, A.T. The Integral Role of Tight Junction Proteins in the Repair of Injured Intestinal Epithelium. Int. J. Mol. Sci. 2020, 21, 972. [Google Scholar] [CrossRef]
- Heinemann, U.; Schuetz, A. Structural Features of Tight-Junction Proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef]
- Park, H.Y.; Kunitake, Y.; Hirasaki, N.; Tanaka, M.; Matsui, T. Theaflavins enhance intestinal barrier of Caco-2 Cell monolayers through the expression of AMP-activated protein kinase-mediated Occludin, Claudin-1, and ZO-1. Biosci. Biotechnol. Biochem. 2015, 79, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Leocádio, P.C.L.; Antunes, M.M.; Teixeira, L.G.; Leonel, A.J.; Alvarez-Leite, J.I.; Machado, D.C.C.; Generoso, S.V.; Cardoso, V.N.; Correia, M.I.T.D. L-Arginine Pretreatment Reduces Intestinal Mucositis as Induced by 5-FU in Mice. Nutr. Cancer 2015, 67, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Youmba, S.B.; Belmonte, L.; Galas, L.; Boukhettala, N.; Bôle-Feysot, C.; Déchelotte, P.; Coëffier, M. Methotrexate Modulates Tight Junctions Through NF-κB, MEK, and JNK Pathways. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 463–470. [Google Scholar] [CrossRef]
- Kanwal, S.; Joseph, T.P.; Owusu, L.; Xiaomeng, R.; Meiqi, L.; Yi, X. A Polysaccharide Isolated from Dictyophora indusiata Promotes Recovery from Antibiotic-Driven Intestinal Dysbiosis and Improves Gut Epithelial Barrier Function in a Mouse Model. Nutrients 2018, 10, 1003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, C. The relationship between intestinal goblet cells and the immune response. Biosci. Rep. 2020, 40, BSR20201471. [Google Scholar] [CrossRef]
- Suzuki, T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 2020, 91, e13357. [Google Scholar] [CrossRef]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef]
- Chen, C.; Chen, S.; Wang, B. A glance at the gut microbiota and the functional roles of the microbes based on marmot fecal samples. Front. Microbiol. 2023, 14, 1035944. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, X.; Fan, Y.; Chen, L.; Ma, X.; Yu, H.; Li, J.; Guan, X.; Zhao, P.; Yang, J. Changes of Intestinal Microbiota in Ovarian Cancer Patients Treated with Surgery and Chemotherapy. Cancer Manag. Res. 2020, 12, 8125–8135. [Google Scholar] [CrossRef]
- Kim, Y.M.; Snijders, A.M.; Brislawn, C.J.; Stratton, K.G.; Zink, E.M.; Fansler, S.J.; Metz, T.O.; Mao, J.H.; Jansson, J.K. Light-Stress Influences the Composition of the Murine Gut Microbiome, Memory Function, and Plasma Metabolome. Front. Mol. Biosci. 2019, 6, 108. [Google Scholar] [CrossRef]
- Andrade, J.C.; Almeida, D.; Domingos, M.; Seabra, C.L.; Machado, D.; Freitas, A.C.; Gomes, A.M. Commensal Obligate Anaerobic Bacteria and Health: Production, Storage, and Delivery Strategies. Front. Bioeng. Biotechnol. 2020, 8, 500. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Yan, J.; Li, M.; Ying, S.; Shi, Z. Gut microbiota dysbiosis increases the risk of visceral gout in goslings through translocation of gut-derived lipopolysaccharide. Poult. Sci. 2019, 98, 5361–5373. [Google Scholar] [CrossRef]
- Sheridan, P.O.; Martin, J.C.; Lawley, T.D.; Browne, H.P.; Harris, H.M.B.; Bernalier-Donadille, A.; Duncan, S.H.; O’Toole, P.W.; Scott, K.P.; Flint, H.J. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb. Genom. 2016, 2, e000043. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Cronin, P.; Joyce, S.A.; O’Toole, P.W.; O’Connor, E.M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef]
- Wong, J.; Piceno, Y.M.; DeSantis, T.Z.; Pahl, M.; Andersen, G.L.; Vaziri, N.D. Expansion of Urease- and Uricase-Containing, Indole- and p-Cresol-Forming and Contraction of Short-Chain Fatty Acid-Producing Intestinal Microbiota in ESRD. Am. J. Nephrol. 2014, 39, 230–237. [Google Scholar] [CrossRef]
- Yamamoto, H.; Ishihara, K.; Takeda, Y.; Koizumi, W.; Ichikawa, T. Changes in the Mucus Barrier during Cisplatin-Induced Intestinal Mucositis in Rats. BioMed Res. Int. 2013, 2013, 276186. [Google Scholar] [CrossRef] [PubMed]











| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | AACGACCCCTTCATTGAC | CCACGACATACTCAGCAC |
| IL-1β | CTCCATGAGCTTTGTACAAGG | TGCTGATGTACCAGTTGGGG |
| IL-6 | TGTGCAATGGCAATTCTGAT | GGTACTCCAGAAGACCAGAGGA |
| TNF-α | CATCTTCTCAAAATTCGAGTGACA | TGGGAGTAGACAAGGTACAACCC |
| Claudin1 | ATCGCAATCTTTGTGTCCACCATT | ATTCTGTTTCCATACCATGCTGTG |
| Occludin | ACTCCTCCAATGGACAAGTG | CCCCACCTGTCGTGTAGTCT |
| ZO-1 | AACCCGAAACTGATGCTATGGA | GCGGCCTTGGAATGTATGTG |
| Mucin-2 | GATGGCACCTACCTCGTTGT | GTCCTGGCACTTGTTGGAAT |
| Control | LBS | CP | CP.LBS | |
|---|---|---|---|---|
| Bacteroidota | 59.33% | 59.81% | 66.22% | 62.64% |
| Firmicutes | 37.78% | 37.81% | 25.46% | 29.98% |
| Proteobacteria | 0.24% | 0.07% | 5.86% | 3.20% |
| Desulfobacterota | 0.57% | 0.99% | 0.65% | 1.90% |
| Actinobacteriota | 0.69% | 0.36% | 1.21% | 0.78% |
| Campilobacterota | 0.96% | 0.35% | 0.19% | 1.14% |
| Cyanobacteria | 0.03% | 0.28% | 0.37% | 0.37% |
| Deferribacterota | 0.67% | 0.15% | 0.06% | 0.04% |
| Patescibacteria | 0.30% | 0.25% | 0.03% | 0.09% |
| Verrucomicrobiota | 0% | 0% | 0% | 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsholi, D.M.; Yacoub, G.S.; Rehman, A.U.; Ullah, H.; Khan, A.I.; Deng, T.; Siddiqui, N.Z.; Alioui, Y.; Farooqui, N.A.; Elkharti, M.; et al. Lactobacillus rhamnosus Attenuates Cisplatin-Induced Intestinal Mucositis in Mice via Modulating the Gut Microbiota and Improving Intestinal Inflammation. Pathogens 2023, 12, 1340. https://doi.org/10.3390/pathogens12111340
Alsholi DM, Yacoub GS, Rehman AU, Ullah H, Khan AI, Deng T, Siddiqui NZ, Alioui Y, Farooqui NA, Elkharti M, et al. Lactobacillus rhamnosus Attenuates Cisplatin-Induced Intestinal Mucositis in Mice via Modulating the Gut Microbiota and Improving Intestinal Inflammation. Pathogens. 2023; 12(11):1340. https://doi.org/10.3390/pathogens12111340
Chicago/Turabian StyleAlsholi, Duaa M., Ghazi Suleiman Yacoub, Ata Ur Rehman, Hidayat Ullah, Asif Iqbal Khan, Ting Deng, Nimra Zafar Siddiqui, Yamina Alioui, Nabeel Ahmed Farooqui, Maroua Elkharti, and et al. 2023. "Lactobacillus rhamnosus Attenuates Cisplatin-Induced Intestinal Mucositis in Mice via Modulating the Gut Microbiota and Improving Intestinal Inflammation" Pathogens 12, no. 11: 1340. https://doi.org/10.3390/pathogens12111340