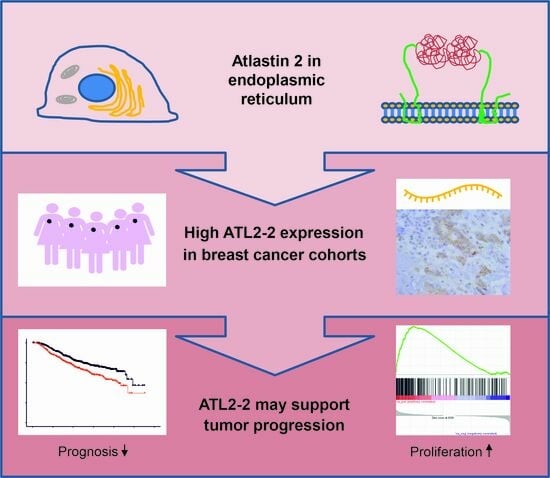
High Atlastin 2-2 (ATL2-2) Expression Associates with Worse Prognosis in Estrogen-Receptor-Positive Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cohorts and Clinical Data
2.2. ATL2-2 mRNA
2.3. ATL2-2 Protein Expression
2.4. Gene Set Enrichment Analysis
2.5. Statistical Analyses
3. Results
3.1. High Expression of ATL2-2 in Breast Tumors
3.2. High Expression of ATL2-2 Associated with Parameters That Indicate Worse Prognosis
3.3. High ATL2-2 mRNA Levels Associated with Shorter Breast-Cancer-Specific Survival
3.4. High ATL2-2 mRNA Associated with Proliferative Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, J.R.; Lingeman, E.; Ahmed, S.; Corn, J.E. Atlastins remodel the endoplasmic reticulum for selective autophagy. J. Cell Biol. 2018, 217, 3354–3367. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Oakes, S.A.; Papa, F.R. The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 2015, 10, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cubillos-Ruiz, J.R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 2021, 21, 71–88. [Google Scholar] [CrossRef]
- Hu, J.; Shibata, Y.; Zhu, P.P.; Voss, C.; Rismanchi, N.; Prinz, W.A.; Rapoport, T.A.; Blackstone, C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 2009, 138, 549–561. [Google Scholar] [CrossRef]
- Orso, G.; Pendin, D.; Liu, S.; Tosetto, J.; Moss, T.J.; Faust, J.E.; Micaroni, M.; Egorova, A.; Martinuzzi, A.; McNew, J.A.; et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 2009, 460, 978–983. [Google Scholar] [CrossRef]
- Jang, E.; Moon, Y.; Yoon, S.Y.; Diaz, J.A.R.; Lee, M.; Ko, N.; Park, J.; Eom, S.H.; Lee, C.; Jun, Y. Human atlastins are sufficient to drive the fusion of liposomes with a physiological lipid composition. J. Cell Biol. 2023, 222, e202109090. [Google Scholar] [CrossRef] [PubMed]
- Rismanchi, N.; Soderblom, C.; Stadler, J.; Zhu, P.P.; Blackstone, C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 2008, 17, 1591–1604. [Google Scholar] [CrossRef]
- Morin-Leisk, J.; Saini, S.G.; Meng, X.; Makhov, A.M.; Zhang, P.; Lee, T.H. An intramolecular salt bridge drives the soluble domain of GTP-bound atlastin into the postfusion conformation. J. Cell Biol. 2011, 195, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Klemm, R.W.; Liu, T.Y.; Zhang, M.; Sun, S.; Sui, X.; Liu, X.; Rapoport, T.A.; Hu, J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 3976–3981. [Google Scholar] [CrossRef]
- Byrnes, L.J.; Sondermann, H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA 2011, 108, 2216–2221. [Google Scholar] [CrossRef]
- Wang, S.; Tukachinsky, H.; Romano, F.B.; Rapoport, T.A. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife 2016, 5, e18605. [Google Scholar] [CrossRef] [PubMed]
- Anggrandariyanny, P.C.; Kajiho, H.; Yamamoto, Y.; Sakisaka, T. Lunapark ubiquitinates atlastin-2 for the tubular network formation of the endoplasmic reticulum. J. Biochem. 2022, 172, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhu, P.P.; Renvoise, B.; Maldonado-Baez, L.; Park, S.H.; Blackstone, C. Mammalian knock out cells reveal prominent roles for atlastin GTPases in ER network morphology. Exp. Cell Res. 2016, 349, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhao, H.; Zhao, Y.G.; Hu, J.; Zhang, H. Atlastin 2/3 regulate ER targeting of the ULK1 complex to initiate autophagy. J. Cell Biol. 2021, 220, e202012091. [Google Scholar] [CrossRef] [PubMed]
- Le Guillou, S.; Laubier, J.; Pechoux, C.; Aujean, E.; Castille, J.; Leroux, C.; Le Provost, F. Defects of the endoplasmic reticulum and changes to lipid droplet size in mammary epithelial cells due to miR-30b-5p overexpression are correlated to a reduction in Atlastin 2 expression. Biochem. Biophys. Res. Commun. 2019, 512, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A Promising Regulator in Development and Disease. Biomed. Res. Int. 2018, 2018, 9623412. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, K.; Li, J.; Liu, Y.; Zhang, Z.; Meng, D. MiR-30e-5p deficiency exerts an inhibitory effect on inflammation in rheumatoid arthritis via regulating Atl2 expression. Arch. Rheumatol. 2023, 38, 119–128. [Google Scholar] [CrossRef]
- Behrendt, L.; Kurth, I.; Kaether, C. A disease causing ATLASTIN 3 mutation affects multiple endoplasmic reticulum-related pathways. Cell. Mol. Life Sci. 2019, 76, 1433–1445. [Google Scholar] [CrossRef]
- McCorquodale, D.S., 3rd; Ozomaro, U.; Huang, J.; Montenegro, G.; Kushman, A.; Citrigno, L.; Price, J.; Speziani, F.; Pericak-Vance, M.A.; Zuchner, S. Mutation screening of spastin, atlastin, and REEP1 in hereditary spastic paraplegia. Clin. Genet. 2011, 79, 523–530. [Google Scholar] [CrossRef]
- Han, J.; Park, H.; Maharana, C.; Gwon, A.R.; Park, J.; Baek, S.H.; Bae, H.G.; Cho, Y.; Kim, H.K.; Sul, J.H.; et al. Alzheimer’s disease-causing presenilin-1 mutations have deleterious effects on mitochondrial function. Theranostics 2021, 11, 8855–8873. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Breast Cancer. WHO News Room. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 12 July 2023).
- Xie, J.; Zou, Y.; Gao, T.; Xie, L.; Tan, D.; Xie, X. Therapeutic Landscape of Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer. Cancer Control 2022, 29, 10732748221099230. [Google Scholar] [CrossRef] [PubMed]
- Blakely, B.; Shin, S.; Jin, K. Overview of the therapeutic strategies for ER positive breast cancer. Biochem. Pharmacol. 2023, 212, 115552. [Google Scholar] [CrossRef] [PubMed]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Munir, A.; Holt, S.; Huws, A.M.; Khan, S.; Davies, D.M.; Khawaja, S.; Sharaiha, Y. A 10 year service evaluation of the survival of 439 patients with early oestrogen receptor positive breast cancer who underwent initial OncotypeDX (R) testing to guide adjuvant chemotherapy decisions. Cancer Treat. Res. Commun. 2023, 34, 100671. [Google Scholar] [CrossRef]
- Savci-Heijink, C.D.; Halfwerk, H.; Koster, J.; van de Vijver, M.J. A novel gene expression signature for bone metastasis in breast carcinomas. Breast Cancer Res. Treat. 2016, 156, 249–259. [Google Scholar] [CrossRef]
- Yu, M.; Huo, S.; Sun, L.; Gao, J.; Liu, Y.; Yu, J.; Liu, F.; Sheng, S.; Nie, X.; Nan, Q.; et al. Epidermal Growth Factor Receptor Mutation Mechanisms in Nonsmall Cell Lung Cancer by Transcriptome Sequencing. Cancer Biother. Radiopharm. 2022, 37, 560–568. [Google Scholar] [CrossRef]
- Zhong, S.; Wang, J.; Hou, J.; Zhang, Q.; Xu, H.; Hu, J.; Zhao, J.; Feng, J. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics 2018, 10, 1301–1313. [Google Scholar] [CrossRef]
- Freiberger, S.N.; Brada, M.; Fritz, C.; Holler, S.; Vogetseder, A.; Horcic, M.; Bihl, M.; Michal, M.; Lanzer, M.; Wartenberg, M.; et al. SalvGlandDx—A comprehensive salivary gland neoplasm specific next generation sequencing panel to facilitate diagnosis and identify therapeutic targets. Neoplasia 2021, 23, 473–487. [Google Scholar] [CrossRef]
- Amirfallah, A.; Arason, A.; Einarsson, H.; Gudmundsdottir, E.T.; Freysteinsdottir, E.S.; Olafsdottir, K.A.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. High expression of the vacuole membrane protein 1 (VMP1) is a potential marker of poor prognosis in HER2 positive breast cancer. PLoS ONE 2019, 14, e0221413. [Google Scholar] [CrossRef]
- Gudmundsdottir, E.T.; Barkardottir, R.B.; Arason, A.; Gunnarsson, H.; Amundadottir, L.T.; Agnarsson, B.A.; Johannsson, O.T.; Reynisdottir, I. The risk allele of SNP rs3803662 and the mRNA level of its closest genes TOX3 and LOC643714 predict adverse outcome for breast cancer patients. BMC Cancer 2012, 12, 621. [Google Scholar] [CrossRef] [PubMed]
- Amirfallah, A.; Knutsdottir, H.; Arason, A.; Hilmarsdottir, B.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. Hsa-miR-21-3p associates with breast cancer patient survival and targets genes in tumor suppressive pathways. PLoS ONE 2021, 16, e0260327. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Rueda, O.M.; Sammut, S.J.; Seoane, J.A.; Chin, S.F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef]
- Crosby, D.; Mikolaj, M.R.; Nyenhuis, S.B.; Bryce, S.; Hinshaw, J.E.; Lee, T.H. Reconstitution of human atlastin fusion activity reveals autoinhibition by the C terminus. J. Cell Biol. 2022, 221, e202107070. [Google Scholar] [CrossRef]
- Xenabrowser. Available online: https://xenabrowser.net/ (accessed on 26 May 2021).
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- GSEA: Gene Set Enrichment Analysis. Available online: https://www.gsea-msigdb.org/gsea/index.jsp (accessed on 22 November 2022).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Chen, K.; Navin, N.E.; Wang, Y.; Schmidt, H.K.; Wallis, J.W.; Niu, B.; Fan, X.; Zhao, H.; McLellan, M.D.; Hoadley, K.A.; et al. BreakTrans: Uncovering the genomic architecture of gene fusions. Genome Biol. 2013, 14, R87. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Enrichr. Available online: https://maayanlab.cloud/Enrichr/ (accessed on 24 November 2022).
- The R Project for Statistical Computing. Available online: http://www.r-project.org (accessed on 24 November 2022).
- Tapial, J.; Ha, K.C.H.; Sterne-Weiler, T.; Gohr, A.; Braunschweig, U.; Hermoso-Pulido, A.; Quesnel-Vallieres, M.; Permanyer, J.; Sodaei, R.; Marquez, Y.; et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017, 27, 1759–1768. [Google Scholar] [CrossRef]
- UCSC Genome Browser. Available online: http://genome.ucsc.edu/ (accessed on 28 May 2021).
- Mochida, K.; Nakatogawa, H. ER-phagy: Selective autophagy of the endoplasmic reticulum. EMBO Rep. 2022, 23, e55192. [Google Scholar] [CrossRef]
- Anderson, C.M.; Macleod, K.F. Autophagy and cancer cell metabolism. Int. Rev. Cell Mol. Biol. 2019, 347, 145–190. [Google Scholar] [CrossRef]
- Wu, L.; Lian, W.; Zhao, L. Calcium signaling in cancer progression and therapy. FEBS J. 2021, 288, 6187–6205. [Google Scholar] [CrossRef]
- Ramirez, O.A.; Cordova, A.; Cerda, M.; Lobos, P.; Hartel, S.; Couve, A.; Hidalgo, C. Ryanodine receptor-mediated Ca(2+) release and atlastin-2 GTPase activity contribute to IP(3)-induced dendritic Ca(2+) signals in primary hippocampal neurons. Cell Calcium 2021, 96, 102399. [Google Scholar] [CrossRef]
- Kelly, C.M.; Zeiger, P.J.; Narayanan, V.; Ramsey, K.; Sondermann, H. A novel insertion mutation in atlastin 1 is associated with spastic quadriplegia, increased membrane tethering, and aberrant conformational switching. J. Biol. Chem. 2022, 298, 101438. [Google Scholar] [CrossRef] [PubMed]




| HR | CI | p-Value | |
|---|---|---|---|
| High ATL2-2 | 1.535 | 1.228–1.918 | 1.6 × 10−4 |
| +age at diagnosis 1 | 1.550 | 1.240–1.937 | 1.2 × 10−4 |
| +grade | 1.429 | 1.139–1.794 | 2.1 × 10−3 |
| +tumor size 1 | 1.436 | 1.147–1.799 | 1.6 × 10−3 |
| +nodes | 1.559 | 1.248–1.949 | 9.4 × 10−5 |
| +stage | 1.473 | 1.144–1.898 | 2.7 × 10−3 |
| +molecular subtype | 1.334 | 1.063–1.673 | 1.3 × 10−2 |
| +HER2 | 1.512 | 1.210–1.890 | 2.8 × 10−4 |
| +Progesterone receptor | 1.480 | 1.183–1.851 | 6.0 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynisdottir, I.; Arason, A.; Freysteinsdottir, E.S.; Kristjansdottir, S.B.; Hilmarsdottir, B.; Traustadottir, G.A.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B. High Atlastin 2-2 (ATL2-2) Expression Associates with Worse Prognosis in Estrogen-Receptor-Positive Breast Cancer. Genes 2023, 14, 1559. https://doi.org/10.3390/genes14081559
Reynisdottir I, Arason A, Freysteinsdottir ES, Kristjansdottir SB, Hilmarsdottir B, Traustadottir GA, Johannsson OT, Agnarsson BA, Barkardottir RB. High Atlastin 2-2 (ATL2-2) Expression Associates with Worse Prognosis in Estrogen-Receptor-Positive Breast Cancer. Genes. 2023; 14(8):1559. https://doi.org/10.3390/genes14081559
Chicago/Turabian StyleReynisdottir, Inga, Adalgeir Arason, Edda S. Freysteinsdottir, Sigrun B. Kristjansdottir, Bylgja Hilmarsdottir, Gunnhildur A. Traustadottir, Oskar T. Johannsson, Bjarni A. Agnarsson, and Rosa B. Barkardottir. 2023. "High Atlastin 2-2 (ATL2-2) Expression Associates with Worse Prognosis in Estrogen-Receptor-Positive Breast Cancer" Genes 14, no. 8: 1559. https://doi.org/10.3390/genes14081559