Extracellular Polysaccharide Receptor and Receptor-Binding Proteins of the Rhodobacter capsulatus Bacteriophage-like Gene Transfer Agent RcGTA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions, and Plasmids
2.2. WT R. capsulatus Genome Library Construction and Recipient Capability Complementation of Strain 37b4
2.3. RcGTA Recipient Capability and Adsorption Assays
2.4. Capsule Stain
2.5. Genome Assembly and Alignment Analysis
2.6. BLAST Analysis
2.7. Creation of Overexpression Constructs and Recombinant Fluorescent Protein Purification
2.8. Fluorescence-Based Binding Assay
3. Results
3.1. Comparison of WT SB1003 and 37b4 Genomes, with a Focus on Genes Needed for Recipient Capability
3.2. Genes Conferring RcGTA Binding, Recipient Capability, and the Presence of a Capsule on Strain 37b4
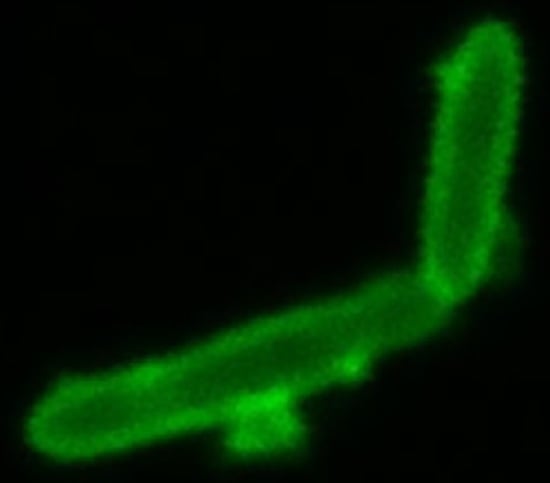
- We also employed Anthony’s stain [32] to visualize the presence or absence of a capsule in microscopy. Figure 7 shows a bright zone due to the capsule around cells of strain SB1003, the absence of such a zone around cells of 37b4, and the presence of a similar, slightly less bright zone around cells of 37b4(pCPS1).
3.3. Binding of Fluorescent Protein-Tagged Tail Fiber and Head Spike Proteins to the WT Strain SB1003, the Quorum-Sensing Mutant ΔgtaI (a gtaI Knockout Lacking a Capsule), 37b4, and 37b4(pCPS1)
- 2.
- The cryo-EM structure of the head spike fiber protein contained high-resolution data of only the N-terminal residues 2 to 10, which showed close interaction (3.5 to 4.9 Å) with residues of the spike base protein pentamer [10]. Therefore, the head spike fiber appears to be anchored by the N terminus, with the bulk of the protein available for interaction with a receptor previously proposed to be the CPS [15]. An AlphaFold model of the head spike fiber protein is given in Figure 10, showing an extended N terminus that interacts with the base protein and a quasi-two domain structure dominated by β strands and loops, as in lectins and other sugar-binding proteins. This AlphaFold structure probably represents only one computationally low-energy-predicted conformation of several structural states. Because the head spike fiber N terminus was found to interact with the base protein pentamer for attachment to the capsid [10], whereas the rest of the protein is thought to bind to a CPS sugar receptor, we designed a synthetic gene encoding a fluorescent protein attached to the N-terminal residue 11, as shown in Supplementary Figure S2.
- 3.
- The fluorescent protein-tagged head spike fiber was used in experiments to evaluate binding to cells. As shown in Figure 11, 100% of this tagged protein bound to the WT strain SB1003 cells, whereas the ΔgtaI mutant (which lacks a capsule) bound about 4%. The 37b4 strain, which also lacks a capsule, bound about 45%, whereas the presence of the pCPS1 cosmid in 37b4 cells increased the binding to about 72%, about 1.6-fold more than the 37b4 strain lacking the cosmid. These results indicate that, as for the tail fiber, the presence of the CPS increases the binding of the head spike fiber. Furthermore, the 37b4 strain bound a percentage of the head spike fiber protein intermediate between the WT strain SB1003 and the cosmid-complemented strain 37b4(pCPS1).
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhaxybayeva, O.; Doolittle, W.F. Lateral gene transfer. Curr. Biol. 2011, 21, R242–R246. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Artzy-Randrup, Y.; Martin, W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 10039–10044. [Google Scholar] [CrossRef] [PubMed]
- Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994, 264, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Biers, E.J.; Wang, K.; Pennington, C.; Belas, R.; Chen, F.; Moran, M.A. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 2008, 74, 2933–2939. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; Deng, L.; Poulos, B.T.; Sullivan, M.B. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ. Microbiol. 2013, 15, 1428–1440. [Google Scholar] [CrossRef]
- Suttle, C. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef]
- McDaniel, L.D.; Young, E.; Delaney, J.; Ruhnau, F.; Ritchie, K.B.; Paul, J.H. High frequency of horizontal gene transfer in the oceans. Science 2010, 330, 50. [Google Scholar] [CrossRef]
- McDaniel, L.D.; Young, E.C.; Ritchie, K.B.; Paul, J.H. Environmental factors influencing gene transfer agent (GTA) mediated transduction in the subtropical ocean. PLoS ONE 2012, 7, e43506. [Google Scholar] [CrossRef]
- Wall, J.D.; Weaver, P.F.; Gest, H. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 1975, 105, 217–224. [Google Scholar] [CrossRef]
- Bárdy, P.; Fuzik, T.; Hrebik, D.; Pantucek, R.; Thomas Beatty, J.; Plevka, P. Structure and mechanism of DNA delivery of a gene transfer agent. Nat. Commun. 2020, 11, 3034. [Google Scholar] [CrossRef]
- Salazar, A.J.; Sherekar, M.; Tsai, J.; Sacchettini, J.C. R pyocin tail fiber structure reveals a receptor-binding domain with a lectin fold. PLoS ONE 2019, 14, e0211432. [Google Scholar] [CrossRef] [PubMed]
- Sycheva, L.V.; Shneider, M.M.; Popova, A.V.; Ziganshin, R.H.; Volozhantsev, N.V.; Miroshnikov, K.A.; Leiman, P.G. Crystal Structure of the putative tail fiber protein gp53 from the Acinetobacter baumannii bacteriophage AP22. bioRxiv 2019, 518761. [Google Scholar] [CrossRef]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents: Phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Brimacombe, C.A.; Stevens, A.; Jun, D.; Mercer, R.; Lang, A.S.; Beatty, J.T. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol. Microbiol. 2013, 87, 802–817. [Google Scholar] [CrossRef] [PubMed]
- Westbye, A.B.; Kuchinski, K.; Yip, C.K.; Beatty, J.T. The Gene Transfer Agent RcGTA Contains Head Spikes Needed for Binding to the Rhodobacter capsulatus Polysaccharide Cell Capsule. J. Mol. Biol. 2016, 428, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Hynes, A.P.; Mercer, R.G.; Watton, D.E.; Buckley, C.B.; Lang, A.S. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol. Microbiol. 2012, 85, 314–325. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Ditta, G.; Schmidhauser, T.; Yakobsen, E.; Lu, P.; Liang, X.-W.; Finlay, D.R.; Guiney, D.; Helinski, D.R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 1985, 13, 149–153. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Marrs, B. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 1974, 71, 971–973. [Google Scholar] [CrossRef]
- Solioz, M.; Marrs, B. The gene transfer agent of Rhodopseudomonas capsulata. Arch. Biochem. Biophys. 1977, 181, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Biebl, H.; Drews, G. Das in-vivo spektrum als taxonomisches merknal bei untersuchungen zur verbreitung von athiorhodacea. Zent. Bakteriol. Parasitenkd. Infekt. Hyg. 1969, 123, 425–452. [Google Scholar]
- Ding, H.; Moksa, M.M.; Hirst, M.; Beatty, J.T. Draft genome sequences of six Rhodobacter capsulatus strains, YW1, YW2, B6, Y262, R121, and DE442. Genome Announc. 2014, 2, e00050-14. [Google Scholar] [CrossRef] [PubMed]
- Beatty, J.T.; Gest, H. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch. Microbiol. 1981, 129, 335–340. [Google Scholar] [CrossRef]
- Friedman, A.M.; Long, S.R.; Brown, S.E.; Buikema, W.J.; Ausubel, F.M. Construction of A Broad Host Range Vector And its Use in the Genetic Analysis of Rhizobium mutants. Gene 1982, 18, 289–296. [Google Scholar] [CrossRef]
- Dastoor, F.; Forrest, M.E.; Beatty, J.T. Cloning, sequencing, and oxygen regulation of the Rhodobacter capsulatus a-ketoglutarate dehydrogenase operon. J. Bacteriol. 1997, 179, 4559–4566. [Google Scholar] [CrossRef]
- Leung, M.M.; Brimacombe, C.A.; Spiegelman, G.B.; Beatty, J.T. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol. Microbiol. 2012, 83, 759–774. [Google Scholar] [CrossRef]
- Anthony, E.E., Jr. A Note on Capsule Staining. Science 1931, 73, 319–320. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Jain, C.; Conrad, R.E.; Aluru, S.; Konstantinidis, K.T. Reply to: “Re-evaluating the evidence for a universal genetic boundary among microbial species”. Nat. Commun. 2021, 12, 4060. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Ding, H.; Beatty, J.T. Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol. Microbiol. 2014, 92, 1260–1278. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Ding, H.; Johnson, J.A.; Beatty, J.T. Homologues of Genetic Transformation DNA Import Genes Are Required for Rhodobacter capsulatus Gene Transfer Agent Recipient Capability Regulated by the Response Regulator CtrA. J. Bacteriol. 2015, 197, 2653–2663. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Koonin, E.V. A Tight Link between Orthologs and Bidirectional Best Hits in Bacterial and Archaeal Genomes. Genome Biol. Evol. 2012, 4, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Hynes, A.P.; Shakya, M.; Mercer, R.G.; Grull, M.P.; Bown, L.; Davidson, F.; Steffen, E.; Matchem, H.; Peach, M.E.; Berger, T.; et al. Functional and Evolutionary Characterization of a Gene Transfer Agent’s Multilocus “Genome”. Mol. Biol. Evol. 2016, 33, 2530–2543. [Google Scholar] [CrossRef] [PubMed]
- Watt, V.M.; Ingles, C.J.; Urdea, M.S.; Rutter, W.J. Homology requirements for recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 1985, 82, 4768–4772. [Google Scholar] [CrossRef]
- Shen, P.; Huang, H.V. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics 1986, 112, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.L.; Taylor, T.A.; Beatty, J.T.; Greenberg, E.P. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 2002, 184, 6515–6521. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]











| SB1003 Blast Query | Gene Designations | Predicted Function [Reference] | Presence in 37b4 |
|---|---|---|---|
| rcc01081 to rcc01086 | rcc1083 = lspL1 | CPS biosynthesis gene cluster [14] | Absent |
| rcc01932 | N/A | CPS-related; repeat unit chain initiation [14] | Present |
| rcc01958 to rcc01960 | wzc, wzb, wza | CPS-related; polysaccharide export [14] | Present |
| rcc00197 | comF | Competence protein F; required for DNA transport from the periplasm to cytoplasm [41] | Present |
| rcc00460 | comM | Competence protein M; putative ATPase and helicase-like domains [41] | Present |
| rcc00222 | radC | DNA repair protein RadC; expressed during competence [41] | Present |
| rcc03098 | dprA | DNA protecting protein A; transformation-dedicated RecA loader, also involved in competence shut-off [41] | Present |
| rcc01751 | recA | Recombination protein A; RecA; DNA repair and homologous recombination [42] | Present |
| Strain | % Mismatch in 200 bp 5′ | % Mismatch in 200 bp 3′ | % Mismatch in 400 bp 5′ | % Mismatch in 400 bp 3′ |
|---|---|---|---|---|
| SB1003 | 0 | 1 | 0 | 0.5 |
| 37b4 | 6.5 | 7 | 8.2 | 6.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alim, N.T.B.; Koppenhöfer, S.; Lang, A.S.; Beatty, J.T. Extracellular Polysaccharide Receptor and Receptor-Binding Proteins of the Rhodobacter capsulatus Bacteriophage-like Gene Transfer Agent RcGTA. Genes 2023, 14, 1124. https://doi.org/10.3390/genes14051124
Alim NTB, Koppenhöfer S, Lang AS, Beatty JT. Extracellular Polysaccharide Receptor and Receptor-Binding Proteins of the Rhodobacter capsulatus Bacteriophage-like Gene Transfer Agent RcGTA. Genes. 2023; 14(5):1124. https://doi.org/10.3390/genes14051124
Chicago/Turabian StyleAlim, Nawshin T. B., Sonja Koppenhöfer, Andrew S. Lang, and J. Thomas Beatty. 2023. "Extracellular Polysaccharide Receptor and Receptor-Binding Proteins of the Rhodobacter capsulatus Bacteriophage-like Gene Transfer Agent RcGTA" Genes 14, no. 5: 1124. https://doi.org/10.3390/genes14051124