A Glimpse into Chromatin Organization and Nuclear Lamina Contribution in Neuronal Differentiation
Abstract
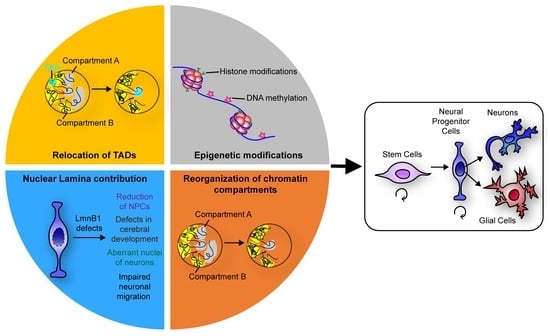
:1. Introduction
2. Three-Dimensional Organization of Chromatin within the Nucleus
3. Nuclear Envelope: LINC Complex and Nuclear Lamins
4. Neurogenesis: An Overview
5. Chromatin Structure Involvement in Neural Development
6. Implication of Nuclear Lamina in Neuronal Development
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allis, C.D.; Jenuwein, T. The Molecular Hallmarks of Epigenetic Control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Useche, I.; Nurse, N.P.; Tian, Y.; Kansara, B.S.; Shim, D.; Yuan, C. DNA Methylation Effects on Tetra-Nucleosome Compaction and Aggregation. Biophys. J. 2014, 107, 1629–1636. [Google Scholar] [CrossRef] [PubMed]
- Choy, J.S.; Wei, S.; Lee, J.Y.; Tan, S.; Chu, S.; Lee, T.-H. DNA Methylation Increases Nucleosome Compaction and Rigidity. J. Am. Chem. Soc. 2010, 132, 1782–1783. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Pappalardo, X.G.; Barra, V. Losing DNA Methylation at Repetitive Elements and Breaking Bad. Epigenet. Chromatin 2021, 14, 25. [Google Scholar] [CrossRef]
- Lande-Diner, L.; Zhang, J.; Ben-Porath, I.; Amariglio, N.; Keshet, I.; Hecht, M.; Azuara, V.; Fisher, A.G.; Rechavi, G.; Cedar, H. Role of DNA Methylation in Stable Gene Repression. J. Biol. Chem. 2007, 282, 12194–12200. [Google Scholar] [CrossRef]
- Wan, J.; Su, Y.; Song, Q.; Tung, B.; Oyinlade, O.; Liu, S.; Ying, M.; Ming, G.-L.; Song, H.; Qian, J.; et al. Methylated Cis-Regulatory Elements Mediate KLF4-Dependent Gene Transactivation and Cell Migration. eLife 2017, 6, e20068. [Google Scholar] [CrossRef]
- Domcke, S.; Bardet, A.F.; Adrian Ginno, P.; Hartl, D.; Burger, L.; Schübeler, D. Competition between DNA Methylation and Transcription Factors Determines Binding of NRF1. Nature 2015, 528, 575–579. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Specificity of the HP1 Chromo Domain for the Methylated N-terminus of Histone H3. EMBO J. 2001, 20, 5232–5241. Available online: https://www.embopress.org/doi/full/10.1093/emboj/20.18.5232 (accessed on 22 April 2023). [CrossRef] [PubMed]
- Hassan, A.H.; Prochasson, P.; Neely, K.E.; Galasinski, S.C.; Chandy, M.; Carrozza, M.J.; Workman, J.L. Function and Selectivity of Bromodomains in Anchoring Chromatin-Modifying Complexes to Promoter Nucleosomes. Cell 2002, 111, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of Multiple Distinct Snf2 Subfamilies with Conserved Structural Motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Logie, C.; Bonte, E.; Becker, P.B.; Wade, P.A.; Wolffe, A.P.; Wu, C.; Imbalzano, A.N.; Peterson, C.L. Functional Delineation of Three Groups of the ATP-Dependent Family of Chromatin Remodeling Enzymes. J. Biol. Chem. 2000, 275, 18864–18870. [Google Scholar] [CrossRef]
- Deuring, R.; Fanti, L.; Armstrong, J.A.; Sarte, M.; Papoulas, O.; Prestel, M.; Daubresse, G.; Verardo, M.; Moseley, S.L.; Berloco, M.; et al. The ISWI Chromatin-Remodeling Protein Is Required for Gene Expression and the Maintenance of Higher Order Chromatin Structure in vivo. Mol. Cell 2000, 5, 355–365. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn Transcriptional Overlap, but Not Its LncRNA Products, Induces Imprinted Igf2r Silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long Non-Coding Antisense RNA Controls Uchl1 Translation through an Embedded SINEB2 Repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Carollo, P.S.; Barra, V. Chromatin Epigenetics and Nuclear Lamina Keep the Nucleus in Shape: Examples from Natural and Accelerated Aging. Biol. Cell 2023, 115, 2200023. [Google Scholar] [CrossRef]
- Malhas, A.; Lee, C.F.; Sanders, R.; Saunders, N.J.; Vaux, D.J. Defects in Lamin B1 Expression or Processing Affect Interphase Chromosome Position and Gene Expression. J. Cell Biol. 2007, 176, 593–603. [Google Scholar] [CrossRef]
- Shah, P.P.; Lv, W.; Rhoades, J.H.; Poleshko, A.; Abbey, D.; Caporizzo, M.A.; Linares-Saldana, R.; Heffler, J.G.; Sayed, N.; Thomas, D.; et al. Pathogenic LMNA Variants Disrupt Cardiac Lamina-Chromatin Interactions and de-Repress Alternative Fate Genes. Cell Stem Cell 2021, 28, 938–954.e9. [Google Scholar] [CrossRef]
- Malhas, A.N.; Lee, C.F.; Vaux, D.J. Lamin B1 Controls Oxidative Stress Responses via Oct-1. J. Cell Biol. 2009, 184, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Cellular Mechanotransduction: Putting All the Pieces Together Again. FASEB J. 2006, 20, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Farge, E. Mechanical Induction of Twist in the Drosophila Foregut/Stomodeal Primordium. Curr. Biol. 2003, 13, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Maeshima, K.; Ide, S.; Babokhov, M. Dynamic Chromatin Organization without the 30-Nm Fiber. Curr. Opin. Cell Biol. 2019, 58, 95–104. [Google Scholar] [CrossRef]
- Majumder, P.; Gomez, J.A.; Chadwick, B.P.; Boss, J.M. The Insulator Factor CTCF Controls MHC Class II Gene Expression and Is Required for the Formation of Long-Distance Chromatin Interactions. J. Exp. Med. 2008, 205, 785–798. [Google Scholar] [CrossRef]
- Nativio, R.; Wendt, K.S.; Ito, Y.; Huddleston, J.E.; Uribe-Lewis, S.; Woodfine, K.; Krueger, C.; Reik, W.; Peters, J.-M.; Murrell, A. Cohesin Is Required for Higher-Order Chromatin Conformation at the Imprinted IGF2-H19 Locus. PLoS Genet. 2009, 5, e1000739. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Toshchakov, S.; Parshikov, A.; Georgiev, P. Study of the Functional Interaction between Mcp Insulators from the Drosophila Bithorax Complex: Effects of Insulator Pairing on Enhancer-Promoter Communication. Mol. Cell. Biol. 2007, 27, 3035. [Google Scholar] [CrossRef]
- Sanborn, A.L.; Rao, S.S.P.; Huang, S.-C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin Extrusion Explains Key Features of Loop and Domain Formation in Wild-Type and Engineered Genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef]
- Nichols, M.H.; Corces, V.G. A Tethered-Inchworm Model of SMC DNA Translocation. Nat. Struct. Mol. Biol. 2018, 25, 906–910. [Google Scholar] [CrossRef]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial Partitioning of the Regulatory Landscape of the X-Inactivation Centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural Protein Subclasses Shape 3-D Organization of Genomes during Lineage Commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Symmons, O.; Uslu, V.V.; Tsujimura, T.; Ruf, S.; Nassari, S.; Schwarzer, W.; Ettwiller, L.; Spitz, F. Functional and Topological Characteristics of Mammalian Regulatory Domains. Genome Res. 2014, 24, 390–400. [Google Scholar] [CrossRef]
- Smith, E.M.; Lajoie, B.R.; Jain, G.; Dekker, J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am. J. Hum. Genet. 2016, 98, 185–201. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Jung, I.; Wu, H.; Zhai, Y.; Tang, Y.; et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef]
- Hnisz, D.; Weintraub, A.S.; Day, D.S.; Valton, A.-L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of Proto-Oncogenes by Disruption of Chromosome Neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M.; Dietzel, S.; Müller, S.; Solovei, I.; Fakan, S. Chromosome Territories—A Functional Nuclear Landscape. Curr. Opin. Cell Biol. 2006, 18, 307–316. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, Z.; Fang, Y.; Chen, Y.; Peng, J.; Song, J.; Li, J.; Shi, J.; Zhou, J.-Q.; Zhao, Y. Chromosome Territory Reorganization through Artificial Chromosome Fusion Is Compatible with Cell Fate Determination and Mouse Development. Cell Discov. 2023, 9, 11. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Barra, V.; Chiavetta, R.F.; Titoli, S.; Provenzano, I.M.; Carollo, P.S.; Di Leonardo, A. Specific Irreversible Cell-Cycle Arrest and Depletion of Cancer Cells Obtained by Combining Curcumin and the Flavonoids Quercetin and Fisetin. Genes 2022, 13, 1125. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain Organization of Human Chromosomes Revealed by Mapping of Nuclear Lamina Interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef]
- Starr, D.A.; Han, M. Role of ANC-1 in Tethering Nuclei to the Actin Cytoskeleton. Science 2002, 298, 406–409. [Google Scholar] [CrossRef]
- Zhang, Q.; Skepper, J.N.; Yang, F.; Davies, J.D.; Hegyi, L.; Roberts, R.G.; Weissberg, P.L.; Ellis, J.A.; Shanahan, C.M. Nesprins: A Novel Family of Spectrin-Repeat-Containing Proteins That Localize to the Nuclear Membrane in Multiple Tissues. J. Cell Sci. 2001, 114, 4485–4498. [Google Scholar] [CrossRef]
- Lygerou, Z.; Christophides, G.; Séraphin, B. A Novel Genetic Screen for SnRNP Assembly Factors in Yeast Identifies a Conserved Protein, Sad1p, Also Required for Pre-MRNA Splicing. Mol. Cell. Biol. 1999, 19, 2008–2020. [Google Scholar] [CrossRef]
- Malone, C.J.; Fixsen, W.D.; Horvitz, H.R.; Han, M. UNC-84 Localizes to the Nuclear Envelope and Is Required for Nuclear Migration and Anchoring during C. Elegans Development. Dev. Camb. Engl. 1999, 126, 3171–3181. [Google Scholar] [CrossRef]
- Guarda, A.; Bolognese, F.; Bonapace, I.M.; Badaracco, G. Interaction between the Inner Nuclear Membrane Lamin B Receptor and the Heterochromatic Methyl Binding Protein, MeCP2. Exp. Cell Res. 2009, 315, 1895–1903. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Callebaut, I.; Pezhman, A.; Courvalin, J.-C.; Worman, H.J. Domain-Specific Interactions of Human HP1-Type Chromodomain Proteins and Inner Nuclear Membrane Protein LBR. J. Biol. Chem. 1997, 272, 14983–14989. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.G.; Lee, R.T. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Almassalha, L.M.; Backman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin Histone Modifications and Rigidity Affect Nuclear Morphology Independent of Lamins. Mol. Biol. Cell 2018, 29, 220–233. [Google Scholar] [CrossRef]
- Makhija, E.; Jokhun, D.S.; Shivashankar, G.V. Nuclear Deformability and Telomere Dynamics Are Regulated by Cell Geometric Constraints. Proc. Natl. Acad. Sci. USA 2016, 113, E32–E40. [Google Scholar] [CrossRef]
- Alisafaei, F.; Jokhun, D.S.; Shivashankar, G.V.; Shenoy, V.B. Regulation of Nuclear Architecture, Mechanics, and Nucleocytoplasmic Shuttling of Epigenetic Factors by Cell Geometric Constraints. Proc. Natl. Acad. Sci. USA 2019, 116, 13200–13209. [Google Scholar] [CrossRef]
- Jain, N.; Iyer, K.V.; Kumar, A.; Shivashankar, G.V. Cell Geometric Constraints Induce Modular Gene-Expression Patterns via Redistribution of HDAC3 Regulated by Actomyosin Contractility. Proc. Natl. Acad. Sci. USA 2013, 110, 11349–11354. [Google Scholar] [CrossRef]
- Lombardi, M.L.; Jaalouk, D.E.; Shanahan, C.M.; Burke, B.; Roux, K.J.; Lammerding, J. The Interaction between Nesprins and Sun Proteins at the Nuclear Envelope Is Critical for Force Transmission between the Nucleus and Cytoskeleton. J. Biol. Chem. 2011, 286, 26743–26753. [Google Scholar] [CrossRef]
- Alam, S.G.; Zhang, Q.; Prasad, N.; Li, Y.; Chamala, S.; Kuchibhotla, R.; Kc, B.; Aggarwal, V.; Shrestha, S.; Jones, A.L.; et al. The Mammalian LINC Complex Regulates Genome Transcriptional Responses to Substrate Rigidity. Sci. Rep. 2016, 6, 38063. [Google Scholar] [CrossRef]
- Stiles, J.; Jernigan, T.L. The Basics of Brain Development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef]
- Yao, B.; Christian, K.M.; He, C.; Jin, P.; Ming, G.-L.; Song, H. Epigenetic Mechanisms in Neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Nothof, S.A.; Magdinier, F.; Van-Gils, J. Chromatin Structure and Dynamics: Focus on Neuronal Differentiation and Pathological Implication. Genes 2022, 13, 639. [Google Scholar] [CrossRef] [PubMed]
- Golob, J.L.; Paige, S.L.; Muskheli, V.; Pabon, L.; Murry, C.E. Chromatin Remodeling during Mouse and Human Embryonic Stem Cell Differentiation. Dev. Dyn. 2008, 237, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The Large Fraction of Heterochromatin in Drosophila Neurons Is Bound by Both B-Type Lamin and HP1a. Epigenet. Chromatin 2018, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yamashita, T. Spatial Organization of Genome Architecture in Neuronal Development and Disease. Neurochem. Int. 2018, 119, 49–56. [Google Scholar] [CrossRef]
- Aoto, T.; Saitoh, N.; Ichimura, T.; Niwa, H.; Nakao, M. Nuclear and Chromatin Reorganization in the MHC-Oct3/4 Locus at Developmental Phases of Embryonic Stem Cell Differentiation. Dev. Biol. 2006, 298, 354–367. [Google Scholar] [CrossRef]
- Le Gros, M.A.; Clowney, E.J.; Magklara, A.; Yen, A.; Markenscoff-Papadimitriou, E.; Colquitt, B.; Myllys, M.; Kellis, M.; Lomvardas, S.; Larabell, C.A. Soft X-ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis in vivo. Cell Rep. 2016, 17, 2125–2136. [Google Scholar] [CrossRef]
- Meshorer, E.; Yellajoshula, D.; George, E.; Scambler, P.J.; Brown, D.T.; Misteli, T. Hyperdynamic Plasticity of Chromatin Proteins in Pluripotent Embryonic Stem Cells. Dev. Cell 2006, 10, 105–116. [Google Scholar] [CrossRef]
- Billia, F.; Baskys, A.; Carlen, P.L.; De Boni, U. Rearrangement of Centromeric Satellite DNA in Hippocampal Neurons Exhibiting Long-Term Potentiation. Mol. Brain Res. 1992, 14, 101–108. [Google Scholar] [CrossRef]
- Solovei, I.; Grandi, N.; Knoth, R.; Volk, B.; Cremer, T. Positional Changes of Pericentromeric Heterochromatin and Nucleoli in Postmitotic Purkinje Cells during Murine Cerebellum Development. Cytogenet. Genome Res. 2004, 105, 302–310. [Google Scholar] [CrossRef]
- Solovei, I.; Kreysing, M.; Lanctôt, C.; Kösem, S.; Peichl, L.; Cremer, T.; Guck, J.; Joffe, B. Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell 2009, 137, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Clowney, E.J.; LeGros, M.A.; Mosley, C.P.; Clowney, F.G.; Markenskoff-Papadimitriou, E.C.; Myllys, M.; Barnea, G.; Larabell, C.A.; Lomvardas, S. Nuclear Aggregation of Olfactory Receptor Genes Governs Their Monogenic Expression. Cell 2012, 151, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Kishi, Y.; Kondo, S.; Gotoh, Y. Transcriptional Activation of Mouse Major Satellite Regions during Neuronal Differentiation. Cell Struct. Funct. 2012, 37, 101–110. [Google Scholar] [CrossRef]
- Fraser, J.; Ferrai, C.; Chiariello, A.M.; Schueler, M.; Rito, T.; Laudanno, G.; Barbieri, M.; Moore, B.L.; Kraemer, D.C.A.; Aitken, S.; et al. Hierarchical Folding and Reorganization of Chromosomes Are Linked to Transcriptional Changes in Cellular Differentiation. Mol. Syst. Biol. 2015, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Decker, B.; Liput, M.; Abdellatif, H.; Yergeau, D.; Bae, Y.; Jornet, J.M.; Stachowiak, E.K.; Stachowiak, M.K. Global Genome Conformational Programming during Neuronal Development Is Associated with CTCF and Nuclear FGFR1—The Genome Archipelago Model. Int. J. Mol. Sci. 2020, 22, 347. [Google Scholar] [CrossRef] [PubMed]
- Terranova, C.; Narla, S.T.; Lee, Y.-W.; Bard, J.; Parikh, A.; Stachowiak, E.K.; Tzanakakis, E.S.; Buck, M.J.; Birkaya, B.; Stachowiak, M.K. Global Developmental Gene Programing Involves a Nuclear Form of Fibroblast Growth Factor Receptor-1 (FGFR1). PLoS ONE 2015, 10, e0123380. [Google Scholar] [CrossRef] [PubMed]
- Marshall, O.J.; Brand, A.H. Chromatin State Changes during Neural Development Revealed by in vivo Cell-Type Specific Profiling. Nat. Commun. 2017, 8, 2271. [Google Scholar] [CrossRef]
- Wang, Y.; Shin, J.-Y.; Nakanishi, K.; Homma, S.; Kim, G.J.; Tanji, K.; Joseph, L.C.; Morrow, J.P.; Stewart, C.L.; Dauer, W.T.; et al. Postnatal Development of Mice with Combined Genetic Depletions of Lamin A/C, Emerin and Lamina-Associated Polypeptide 1. Hum. Mol. Genet. 2019, 28, 2486–2500. [Google Scholar] [CrossRef]
- Lochs, S.J.A.; Kefalopoulou, S.; Kind, J. Lamina Associated Domains and Gene Regulation in Development and Cancer. Cells 2019, 8, 271. [Google Scholar] [CrossRef]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.M.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef]
- Hirano, Y.; Hizume, K.; Kimura, H.; Takeyasu, K.; Haraguchi, T.; Hiraoka, Y. Lamin B Receptor Recognizes Specific Modifications of Histone H4 in Heterochromatin Formation. J. Biol. Chem. 2012, 287, 42654–42663. [Google Scholar] [CrossRef]
- Williams, R.R.E.; Azuara, V.; Perry, P.; Sauer, S.; Dvorkina, M.; Jørgensen, H.; Roix, J.; McQueen, P.; Misteli, T.; Merkenschlager, M.; et al. Neural Induction Promotes Large-Scale Chromatin Reorganisation of the Mash1 Locus. J. Cell Sci. 2006, 119, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Coffinier, C.; Jung, H.-J.; Nobumori, C.; Chang, S.; Tu, Y.; Barnes, R.H.; Yoshinaga, Y.; de Jong, P.J.; Vergnes, L.; Reue, K.; et al. Deficiencies in Lamin B1 and Lamin B2 Cause Neurodevelopmental Defects and Distinct Nuclear Shape Abnormalities in Neurons. Mol. Biol. Cell 2011, 22, 4683–4693. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-J.; Nobumori, C.; Goulbourne, C.N.; Tu, Y.; Lee, J.M.; Tatar, A.; Wu, D.; Yoshinaga, Y.; de Jong, P.J.; Coffinier, C.; et al. Farnesylation of Lamin B1 Is Important for Retention of Nuclear Chromatin during Neuronal Migration. Proc. Natl. Acad. Sci. USA 2013, 110, E1923–E1932. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Reguant, L.; Blanco, E.; Galan, S.; Le Dily, F.; Cuartero, Y.; Serra-Bardenys, G.; Di Carlo, V.; Iturbide, A.; Cebrià-Costa, J.P.; Nonell, L.; et al. Lamin B1 Mapping Reveals the Existence of Dynamic and Functional Euchromatin Lamin B1 Domains. Nat. Commun. 2018, 9, 3420. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, J.; Yue, S.; Kristiani, L.; Kim, M.; Sauria, M.; Taylor, J.; Kim, Y.; Zheng, Y. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 2018, 71, 802–815.e7. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Li, M.; Shao, S.; Li, C.; Ai, S.; Xue, B.; Hou, Y.; Zhang, Y.; Li, R.; Fan, X.; et al. Nuclear Peripheral Chromatin-Lamin B1 Interaction Is Required for Global Integrity of Chromatin Architecture and Dynamics in Human Cells. Protein Cell 2022, 13, 258–280. [Google Scholar] [CrossRef]
- Gigante, C.M.; Dibattista, M.; Dong, F.N.; Zheng, X.; Yue, S.; Young, S.G.; Reisert, J.; Zheng, Y.; Zhao, H. Lamin B1 Is Required for Mature Neuron-Specific Gene Expression during Olfactory Sensory Neuron Differentiation. Nat. Commun. 2017, 8, 15098. [Google Scholar] [CrossRef]
- Chen, N.Y.; Yang, Y.; Weston, T.A.; Belling, J.N.; Heizer, P.; Tu, Y.; Kim, P.; Edillo, L.; Jonas, S.J.; Weiss, P.S.; et al. An Absence of Lamin B1 in Migrating Neurons Causes Nuclear Membrane Ruptures and Cell Death. Proc. Natl. Acad. Sci. USA 2019, 116, 25870–25879. [Google Scholar] [CrossRef]
- Razafsky, D.; Ward, C.; Potter, C.; Zhu, W.; Xue, Y.; Kefalov, V.J.; Fong, L.G.; Young, S.G.; Hodzic, D. Lamin B1 and Lamin B2 Are Long-Lived Proteins with Distinct Functions in Retinal Development. Mol. Biol. Cell 2016, 27, 1928–1937. [Google Scholar] [CrossRef]
- Good, K.V.; Vincent, J.B.; Ausió, J. MeCP2: The Genetic Driver of Rett Syndrome Epigenetics. Front. Genet. 2021, 12, 620859. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Kaur, M.; Yaeger, D.; Rampuria, A.; Korolev, S.; Pie, J.; Gil-Rodríguez, C.; Arnedo, M.; Loeys, B.; Kline, A.D.; et al. Mutations in Cohesin Complex Members SMC3 and SMC1A Cause a Mild Variant of Cornelia de Lange Syndrome with Predominant Mental Retardation. Am. J. Hum. Genet. 2007, 80, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Squeo, G.M.; Augello, B.; Massa, V.; Milani, D.; Colombo, E.A.; Mazza, T.; Castellana, S.; Piccione, M.; Maitz, S.; Petracca, A.; et al. Customised Next-Generation Sequencing Multigene Panel to Screen a Large Cohort of Individuals with Chromatin-Related Disorder. J. Med. Genet. 2020, 57, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Papantonis, A.; Cook, P.R. Transcription Factories: Genome Organization and Gene Regulation. Chem. Rev. 2013, 113, 8683–8705. [Google Scholar] [CrossRef]
- Nott, A.; Holtman, I.R.; Coufal, N.G.; Schlachetzki, J.C.M.; Yu, M.; Hu, R.; Han, C.Z.; Pena, M.; Xiao, J.; Wu, Y.; et al. Brain Cell Type-Specific Enhancer-Promoter Interactome Maps and Disease-Risk Association. Science 2019, 366, 1134–1139. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martino, S.; Carollo, P.S.; Barra, V. A Glimpse into Chromatin Organization and Nuclear Lamina Contribution in Neuronal Differentiation. Genes 2023, 14, 1046. https://doi.org/10.3390/genes14051046
Martino S, Carollo PS, Barra V. A Glimpse into Chromatin Organization and Nuclear Lamina Contribution in Neuronal Differentiation. Genes. 2023; 14(5):1046. https://doi.org/10.3390/genes14051046
Chicago/Turabian StyleMartino, Salvatore, Pietro Salvatore Carollo, and Viviana Barra. 2023. "A Glimpse into Chromatin Organization and Nuclear Lamina Contribution in Neuronal Differentiation" Genes 14, no. 5: 1046. https://doi.org/10.3390/genes14051046