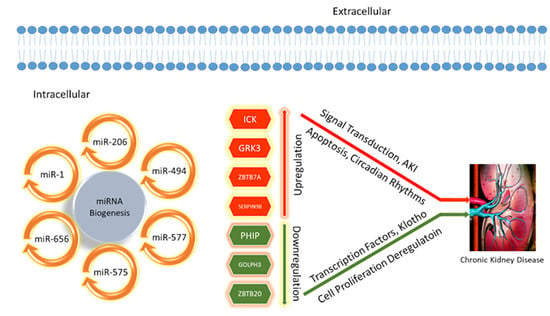
Computational Analysis Reveals Distinctive Interaction of miRNAs with Target Genes in the Pathogenesis of Chronic Kidney Disease
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andrassy, K.M. Comments on ‘KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease’. Kidney Int. 2013, 84, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Pandey, M. Role of kidney biomarkers of chronic kidney disease: An update. Saudi J. Biol. Sci. 2014, 21, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Hostetter, T.H. Progression of renal disease and renal hypertrophy. Annu. Rev. Physiol. 1995, 57, 263–278. [Google Scholar] [CrossRef]
- Pillebout, E.; Weitzman, J.B.; Burtin, M.; Martino, C.; Federici, P.; Yaniv, M.; Friedlander, G.; Terzi1Terzi, F. JunD protects against chronic kidney disease by regulating paracrine mitogens. J. Clin. Investig. 2003, 112, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Jha, V.; Garcia-Garcia, G.; Iseki, K.; Li, Z.; Naicker, S.; Plattner, B.; Saran, R.; Wang, A.Y.-M.; Yang, C.-W. Chronic kidney disease: Global dimension and perspectives. Lancet 2013, 382, 260–272. [Google Scholar] [CrossRef]
- Stumpers, S.; Thomson, N. Review of Kidney Disease and Urologic Disorders among Indigenous People; Australian Indigenous HealthInfoNet: Perth, WA, Australia, 2013; Volume 13, pp. 1–22. [Google Scholar]
- Lea, J.P.; Nicholas, S.B. Diabetes mellitus and hypertension: Key risk factors for kidney disease. J. Natl. Med. Assoc. 2002, 94 (Suppl. S8), 7S–15S. [Google Scholar]
- Gansevoort, R.T.; de Jong, P.E. Challenges for the present CKD classification system. Curr. Opin. Nephrol. Hypertens. 2010, 19, 308–314. [Google Scholar] [CrossRef]
- Pontillo, C.; Zhang, Z.-Y.; Schanstra, J.P.; Jacobs, L.; Zürbig, P.; Thijs, L.; Ramírez-Torres, A.; Heerspink, H.J.; Lindhardt, M.; Klein, R.; et al. Prediction of chronic kidney disease stage 3 by CKD273, a urinary proteomic biomarker. Kidney Int. Rep. 2017, 2, 1066–1075. [Google Scholar] [CrossRef]
- Anders, H.J.; Andersen, K.; Stecher, B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013, 83, 1010–1016. [Google Scholar] [CrossRef]
- Kovesdy, C.P. Epidemiology of chronic kidney disease: An update 2022. Kidney Int. Suppl. 2022, 12, 7–11. [Google Scholar] [CrossRef]
- Tanner, R.M.; Gutiérrez, O.M.; Judd, S.; McClellan, W.; Bowling, C.B.; Bradbury, B.D.; Safford, M.M.; Cushman, M.; Warnock, D.; Muntner, P. Geographic variation in CKD prevalence and ESRD incidence in the United States: Results from the reasons for geographic and racial differences in stroke (REGARDS) study. Am. J. Kidney Dis. 2013, 61, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Brück, K.; Stel, V.S.; Gambaro, G.; Hallan, S.; Völzke, H.; Ärnlöv, J.; Kastarinen, M.; Guessous, I.; Vinhas, J.; Stengel, B.; et al. CKD prevalence varies across the European general population. J. Am. Soc. Nephrol. 2016, 27, 2135–2147. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Prevalence of chronic kidney disease in China–Autho’’s reply. Lancet 2012, 380, 214–216. [Google Scholar] [CrossRef]
- Zdrojewski, Ł.; Zdrojewski, T.; Rutkowski, M.; Bandosz, P.; Król, E.; Wyrzykowski, B.; Rutkowski, B. Prevalence of chronic kidney disease in a representative sample of the Polish population: Results of the NATPOL 2011 survey. Nephrol. Dial. Transplant. 2016, 31, 433–439. [Google Scholar] [CrossRef]
- Cockwell, P.; Fisher, L.A. The global burden of chronic kidney disease. Lancet 2020, 395, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Burrows, N.R.; Vassalotti, J.A.; Saydah, S.H.; Stewart, R.; Gannon, M.; Chen, S.-C.; Li, S.; Pederson, S.; Collins, A.J.; Williams, D.E. Identifying high-risk individuals for chronic kidney disease: Results of the CHERISH Community Demonstration Project. Am. J. Nephrol. 2018, 48, 447–455. [Google Scholar] [CrossRef]
- Imtiaz, S.; Salman, B.; Qureshi, R.; Drohlia, M.F.; Ahmad, A. A review of the epidemiology of chronic kidney disease in Pakistan: A global and regional perspective. Saudi J. Kidney Dis. Transpl. 2018, 29, 1441. [Google Scholar] [CrossRef]
- Mallappallil, M.; Friedman, E.A.; Delano, B.G.; McFarlane, S.I.; Salifu, M.O. Chronic kidney disease in the elderly: Evaluation and management. Clin. Pract. 2014, 11, 525. [Google Scholar] [CrossRef]
- Liang, M.; Liu, Y.; Mladinov, D.; Cowley, A.W., Jr.; Trivedi, H.; Fang, Y.; Xu, X.; Ding, X.; Tian, Z. MicroRNA: A new frontier in kidney and blood pressure research. Am. J. Physiol. Renal Physiol. 2009, 297, F553–F558. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Dandare, A.; Khan, M.J.; Naeem, A.; Liaquat, A. Clinical relevance of circulating non-coding RNAs in metabolic diseases: Emphasis on obesity, diabetes, cardiovascular diseases and metabolic syndrome. Genes Dis. 2022, in press. [Google Scholar] [CrossRef]
- Khan, A.; Andleeb, Z.; Mumtaz, S.; Fatmi, M.Q.; Khan, M.J. Integrated in silico analysis to study the role of microRNAs in the detection of chronic kidney diseases. Curr. Bioinform. 2020, 15, 144–154. [Google Scholar] [CrossRef]
- Conserva, F.; Barozzino, M.; Pesce, F.; Divella, C.; Oranger, A.; Papale, M.; Sallustio, F.; Simone, S.; Laviola, L.; Giorgino, F.; et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of kidney fibrosis in diabetic nephropathy. Sci. Rep. 2019, 9, 11357. [Google Scholar] [CrossRef] [PubMed]
- Trevisani, F.; Ghidini, M.; Larcher, A.; Lampis, A.; Lote, H.; Manunta, P.; Alibrandi, M.T.S.; Zagato, L.; Citterio, L.; Dell’Antonio, G.; et al. MicroRNA 193b-3p as a predictive biomarker of chronic kidney disease in patients undergoing radical nephrectomy for renal cell carcinoma. Br. J. Cancer 2016, 115, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, B.; Jeyamohan, S.; Padmanabhan, G.; Velangann, A.J.; Ramanathan, K. Circulatory microRNA expression profile for coronary artery calcification in chronic kidney disease patients. Afr. Health Sci. 2021, 21, 728–734. [Google Scholar] [CrossRef]
- Scherer, A.; Günther, O.P.; Balshaw, R.F.; Hollander, Z.; Wilson-McManus, J.; Ng, R.; McMaster, W.R.; McManus, B.M.; Keown, P.A. Alteration of human blood cell transcriptome in uremia. BMC Med. Genet. 2013, 6, 23. [Google Scholar] [CrossRef]
- Al-Chaqmaqchi, H.A.M.; Moshfegh, A.; Dadfar, E.; Paulsson, J.; Hassan, M.; Jacobson, S.H.; Lundahl, J. Activation of Wnt/β-catenin pathway in monocytes derived from chronic kidney disease patients. PLoS ONE 2013, 8, e68937. [Google Scholar] [CrossRef]
- Nakagawa, S.; Nishihara, K.; Miyata, H.; Shinke, H.; Tomita, E.; Kajiwara, M.; Matsubara, T.; Iehara, N.; Igarashi, Y.; Yamada, H.; et al. Molecular markers of tubulointerstitial fibrosis and tubular cell damage in patients with chronic kidney disease. PLoS ONE 2015, 10, e0136994. [Google Scholar] [CrossRef]
- Sur, S.; Nguyen, M.; Boada, P.; Sigdel, T.K.; Sollinger, H.; Sarwal, M.M. FcER1: A Novel Molecule Implicated in the Progression of Human Diabetic Kidney Disease. Front. Immunol. 2021, 12, 769972. [Google Scholar] [CrossRef]
- Dandare, A.; Rabia, G.; Khan, M.J. In Silico Analysis of Non-Coding RNAs and Putative Target Genes Implicated in Metabolic Syndrome. Comput. Biol. Med. 2021, 130, 104229. [Google Scholar] [CrossRef]
- Rafiq, M.; Dandare, A.; Javed, A.; Liaquat, A.; Raja, A.A.; Awan, H.M.; Khan, M.J.; Naeem, A. Competing Endogenous RNA Regulatory Networks ofhasa_circ_0126672 in Pathophysiology of Coronary Heart Disease. Genes 2023, 14, 550. [Google Scholar] [CrossRef]
- Iqbal, S.; Ejaz, H.; Nawaz, M.S.; Loor, J.J.; Naeem, A. Meta-analysis of cancer transcriptomes: A new approach to uncover molecular pathological events in different cancer tissues. Network Biol. 2014, 4, 1–20. [Google Scholar]
- Naeem, A.; Zhong, K.; Moisá, S.J.; Drackley, J.K.; Moyes, K.M.; Loor, J.J. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J. Dairy Sci. 2012, 95, 6397–6408. [Google Scholar] [CrossRef] [PubMed]
- Rudnicki, M.; Perco, P.; D′haene, B.; Leierer, J.; Heinzel, A.; Mühlberger, I.; Schweibert, N.; Sunzenauer, J.; Regele, H.; Kronbichler, A.; et al. Renal micro RNA-and RNA-profiles in progressive chronic kidney disease. Eur. J. Clin. Investig. 2016, 46, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Mi, Q.S.; Dong, Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life 2013, 65, 602–614. [Google Scholar] [CrossRef]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Tang, C.; Livingston, M.J.; Liu, Z.; Dong, Z. Autophagy in kidney homeostasis and disease. Nat. Rev. Nephrol. 2020, 16, 489–508. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef]
- Shao, L.; Ma, Y.; Fang, Q.; Huang, Z.; Wan, S.; Wang, J.; Yang, L. Role of protein phosphatase 2A in kidney disease. Exp. Ther. Med. 2021, 22, 1236. [Google Scholar] [CrossRef]
- Helin, K.; Minucci, S. The role of chromatin-associated proteins in cancer. Annu. Rev. Cancer Biol. 2017, 1, 355–377. [Google Scholar] [CrossRef]
- Ruiz-Andres, O.; Sanchez-Niño, M.D.; Moreno, J.A.; Ruiz-Ortega, M.; Ramos, A.M.; Sanz, A.B.; Ortiz, A. Downregulation of kidney protective factors by inflammation: Role of transcription factors and epigenetic mechanisms. Am. J. Physiol. Renal Physiol. 2016, 311, F1329–F1340. [Google Scholar] [CrossRef] [PubMed]
- Canaud, G.; Bonventre, J.V. Cell cycle arrest and the evolution of chronic kidney disease from acute kidney injury. Nephrol. Dial. Transplant. 2015, 30, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Tchkonia, T.; Zhu, Y.; Van Deursen, J.; Campisi, J.; Kirkland, J.L. Cellular senescence and the senescent secretory phenotype: Therapeutic opportunities. J. Clin. Investig. 2013, 123, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Schnaper, H.W.; Jandeska, S.; Runyan, C.E.; Hubchak, S.C.; Basu, R.K.; Curley, J.F.; Smith, R.D.; Hayashida, T. TGF-β signal transduction in chronic kidney disease. Front. Biosci. (Landmark Ed.) 2009, 14, 2448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bao, S.; Wang, D.; Lu, W.; Xu, S.; Zhou, W.; Wang, X.; Xu, X.; Ding, X.; Zhao, S. Downregulation of KLF10 contributes to the regeneration of survived renal tubular cells in cisplatin-induced acute kidney injury via ZBTB7A-KLF10-PTEN axis. Cell Death Discov. 2023, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Guo, J.; Moledina, D.G.; Cantley, L.G. Immune-mediated tubule atrophy promotes acute kidney injury to chronic kidney disease transition. Nat. Commun. 2022, 13, 4892. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wei, L.; Chen, J.; Chen, Z. LncRNA NORAD deficiency alleviates kidney injury in mice and decreases the inflammatory response and apoptosis of lipopolysaccharide-stimulated HK-2 cells via the miR-577/GOLPH3 axis. Cytokine 2022, 153, 155844. [Google Scholar] [CrossRef]
- Zhou, D.; Fu, H.; Zhang, L.; Zhang, K.; Min, Y.; Xiao, L.; Lin, L.; Bastacky, S.I.; Liu, Y. Tubule-derived Wnts are required for fibroblast activation and kidney fibrosis. J. Am. Soc. Nephrol. 2017, 28, 2322–2336. [Google Scholar] [CrossRef]
- Zhou, D.; Fu, H.; Zhang, L.; Zhang, K.; Min, Y.; Xiao, L.; Lin, L.; Bastacky, S.I.; Liu, Y. Wnt3a: Functions and implications in cancer. Chin. J. Cancer 2015, 34, 1–9. [Google Scholar] [CrossRef]
- Kiewisz, J.; Skowronska, A.; Winiarska, A.; Pawlowska, A.; Kiezun, J.; Rozicka, A.; Perkowska-Ptasinska, A.; Kmiec, Z.; Stompor, T. WNT4 expression in primary and secondary kidney diseases: Dependence on staging. Kidney Blood Press. Res. 2019, 44, 200–210. [Google Scholar] [CrossRef]
- Li, B.; Rauhauser, A.A.; Dai, J.; Sakthivel, R.; Igarashi, P.; Jetten, A.M.; Attanasio, M. Increased hedgehog signaling in postnatal kidney results in aberrant activation of nephron developmental programs. Hum. Mol. Genet. 2011, 20, 4155–4166. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, C.M.; Crouthamel, M.H.; Kapustin, A.; Giachelli, C.M. Arterial calcification in chronic kidney disease: Key roles for calcium and phosphate. Circ. Res. 2011, 109, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Olszak, I.T.; Poznansky, M.C.; Evans, R.H.; Olson, D.; Kos, C.; Pollak, M.R.; Brown, E.M.; Scadden, D.T. Extracellular calcium elicits a chemokinetic response from monocytes in vitro and in vivo. J. Clin. Investig. 2000, 105, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Renkema, K.Y.; Alexander, R.T.; Bindels, R.J.; Hoenderop, J.G. Calcium and phosphate homeostasis: Concerted interplay of new regulators. Ann. Med. 2008, 40, 82–91. [Google Scholar] [CrossRef]







| miRNAs | Number of Target DEGs |
|---|---|
| hsa-miR-1 | 117 |
| hsa-miR-206 | 118 |
| hsa-miR-494 | 204 |
| hsa-miR-575 | 40 |
| hsa-miR-577 | 127 |
| hsa-miR-656 | 149 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salim, H.M.U.; Dandare, A.; Khalil, F.; Liaquat, A.; Khan, M.J.; Naeem, A. Computational Analysis Reveals Distinctive Interaction of miRNAs with Target Genes in the Pathogenesis of Chronic Kidney Disease. Genes 2023, 14, 898. https://doi.org/10.3390/genes14040898
Salim HMU, Dandare A, Khalil F, Liaquat A, Khan MJ, Naeem A. Computational Analysis Reveals Distinctive Interaction of miRNAs with Target Genes in the Pathogenesis of Chronic Kidney Disease. Genes. 2023; 14(4):898. https://doi.org/10.3390/genes14040898
Chicago/Turabian StyleSalim, Hafiz Muhammad Umar, Abdullahi Dandare, Fareeha Khalil, Afrose Liaquat, Muhammad Jawad Khan, and Aisha Naeem. 2023. "Computational Analysis Reveals Distinctive Interaction of miRNAs with Target Genes in the Pathogenesis of Chronic Kidney Disease" Genes 14, no. 4: 898. https://doi.org/10.3390/genes14040898