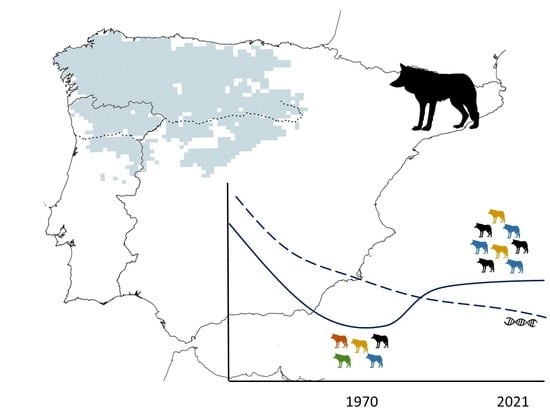
Loss of Mitochondrial Genetic Diversity despite Population Growth: The Legacy of Past Wolf Population Declines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Data

| Sample | Collection # | Year | Locality | Coverage | CR | MT |
|---|---|---|---|---|---|---|
| JAL7556 | EBD 7861M | 1972 | Spain, Extremadura, Badajoz, Villanueva del Fresno | 3.2 | lu1 | MT1 |
| JAL7533 | EBD 7845M | 1972 | Spain, Asturias, Allande | 3.8 | lu4 | MT6 |
| JAL7555 | EBD 7835M | c1970 | Spain, Andalucía, Huelva, Puebla de Guzmán | 35.8 | lu1 | MT4 |
| JAL7559 | EBD 18003M | 1977 | Spain, Andalucía, Jaén, Andújar | 48.9 | lu1 | MT4 |
| JAL7562 | EBD 15858M | 1979 | Spain, Castilla y León, Salamanca, Hinojosa del Duero | 153.4 | lu1 | MT7 |
| JAL7563 | EBD 15039M | 1974 | Spain, Galicia, La Coruña, Monte Xalo | 29.6 | lu2 | MT2 |
| JAL7567 | EBD 15139M | 1975 | Spain, Castilla y León, Palencia, Herrera de Pisuerga | 15.8 | lu4 | MT6 |
| JAL7573 | EBD 7826M | 1979 | Portugal, Guarda, Beiras y Serra da Estrela, Ciudad Rodrigo | 108.7 | lu1 | MT3 |
| JAL7574 | EBD 15865M | c1970 | Spain, Asturias, Cangas de Narcea | 141.4 | lu2 | MT2 |
| JAL7576 | EBD 14871M | 1974 | Spain, Castilla y León, Zamora, Ferreruela | 3.4 | lu1 | MT13 |
| JAL7578 | EBD 15880M | 1971 | Spain, Castilla y León, Burgos, Sedano | 8.1 | lu4 | MT6 |
| JAL7581 | EBD 7888M | 1974 | Spain, Castilla y León, León, Puente de Sanabria | 5.9 | lu1 | MT12 |
| JAL7583 | EBD 14856M | 1974 | Spain, Galicia, Orense, Nogueira de Ramuin. Cortecadela | 0.4 | - | MT8 |
| JAL7585 | EBD 14868M | 1974 | Spain, Galicia, Orense, La Gudiña | 1.9 | - | MT3 |
| JAL7586 | EBD 15893M | 1975 | Spain, Castilla y León, Burgos, Ordejon de Abajo O Santa Maria-humada | 16 | lu4 | MT11 |
| JAL7587 | EBD 7534M | 1984 | Spain, Castilla y León, Palencia, Becerril de Campos | 46.1 | lu4 | MT6 |
| MW916084 * | Cl-H10 | 1975 | Spain | - | lu1 | MT1 |
| MW916085 * | Cl-H117 | 1970 | Spain | - | lu1 | MT4 |
| MW916086 * | Cl-H119 | 1944 | Spain | - | lu1 | MT4 |
| Sample | Locality | CR | MT | Ref. |
|---|---|---|---|---|
| CVA554 | Spain, Castilla y León, Segovia, Cerezo de Arriba | lu4 | MT6 | † |
| CVA558 | Spain, Castilla y León, Palencia, Valdespina | lu4 | MT9 | † |
| CVA560 | Spain, Castilla y León, Zamora, Padornelo | lu2 | MT2 | † |
| CVA564 | Spain, Castilla y León, León, Valverde Enrique | lu1 | MT1 | † |
| CVA568 | Spain, Castilla y León, Zamora, Tábara | lu1 | MT3 | † |
| CVA609 | Spain, Castilla y León, Salamanca, San Cristóbal del Monte | lu1 | MT1 | † |
| JAL7481 | Spain, Castilla-La Mancha, Guadalajara, Pinilla de Jadraque (5506-1) | lu1 | MT1 | † |
| JAL7487 | Spain, Castilla y León, Zamora, Pereruela de Sayago (S Duero) | lu2 | MT2 | † |
| JAL7489 | Spain, Castilla y León, Burgos, Berberana | lu1 | MT1 | † |
| JAL7490 | Spain, Galicia, A Coruña, Ordes (C-1117-2019) | lu2 | MT2 | † |
| JAL7494 | Spain, Galicia, Ourense, Rairiz de Veiga (OU-251-20202) | lu1 | MT8 | † |
| JAL7496 | Spain, Galicia, Lugo, Lugo (V-9326) | lu2 | MT2 | † |
| JAL7599 | Spain, Cantabria, Cabuérniga (ELC-20-33) | lu4 | MT10 | † |
| JAL7604 | Spain, Asturias, Tineo (EBD 33175M) | lu2 | MT2 | † |
| JAL7609 | Spain, Asturias, Caso (EBD 33180M) | lu1 | MT1 | † |
| JAL7613 | Spain, Castilla y León, Burgos | lu4 | MT6 | † |
| JAL7617 | Portugal, Serra da Anta, Merufe (SMLM 139) | lu1 | MT8 | † |
| wEEP | Spain, Captive (SRR3384029;SRR3384030) | lu1 | MT1 | [32] |
| wSierraMorena | Spain, Andalucía, Jaén, Andújar (SRR7586076) | lu1 | MT4 | [32] |
| wSpain | Spain (SRR1518528;SRR1518529) | lu4 | MT6 | [51] |
| wPortugal | Portugal (SRR3574846;SRR1518524;SRR1518523) | lu1 | MT8 | [51] |
| MN071205 | Spain, Captive (MN071205) | lu2 | MT2 | [46] |
| MW916087 | Portugal (MW916087) | - | MT2 | [49] |
| DQ480505 | Spain (DQ480505) | lu1 | MT3 | [44] |
| MW916078 | Portugal (MW916078) | lu1 | MT3 | [49] |
| KU644670 | Spain (KU644670) | lu4 | MT6 | [45] |
| KT448278 | Portugal (KT448278) | lu1 | MT8 | [50] |
| KU644668 | Portugal (KU644668) | lu1 | MT8 | [45] |
| MW916079 | Portugal (MW916079) | - | MT8 | [49] |
2.2. Molecular Methods
2.3. Data Analyses
3. Results
| Haplotype | Historical | Modern | |
|---|---|---|---|
| MT | MT1 | 1 | 6 |
| MT2 | 2 | 7 | |
| MT3 | 2 | 3 | |
| MT4 | 4 | 1 | |
| MT6 | 4 | 4 | |
| MT7 | 1 | - | |
| MT8 | 1 | 6 | |
| MT9 | - | 1 | |
| MT10 | - | 1 | |
| MT11 | 1 | - | |
| MT12 | 1 | - | |
| MT13 | 1 | - | |
| n | 19 | 29 | |
| n H | 10 | 8 | |
| H private | 4 | 2 | |
| CR | lu1 | 10 | 15 |
| lu2 | 2 | 6 | |
| lu4 * | 5 | 6 | |
| n | 17 | 27 | |
| n H | 3 | 3 | |
| H private | 0 | 0 |
| CR (425 bp) | MT (15,460 bp) | |||
|---|---|---|---|---|
| Historical | Modern | Historical | Modern | |
| n | 17 | 27 | 19 | 29 |
| H | 3 | 3 | 10 | 8 |
| S | 6 | 6 | 41 | 39 |
| π | 5.71 × 10−3 (3.6 × 10−3) | 5.07 × 10−3 (3.2 × 10−3) | 9.4 × 10−4 (4.9 × 10−4) | 8.2 × 10−4 (4.3 × 10−4) |
| Hd | 0.588 (0.091) | 0.615 (0.068) | 0.912 (0.036) | 0.852 (0.030) |
| ΘS | 1.78 (0.92) | 1.56 (0.78) | 11.73 (4.34) | 9.93 (3.41) |
| Θπ | 2.43 (1.55) | 2.15 (1.37) | 12.83 (6.76) | 12.74 (6.58) |
| ΘK | 0.77 [0.22, 2.46] | 0.64 [0.19, 1.96] | 10.05 [4.29, 23.67] | 3.29 [1.44, 7.23] |



4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Sotnikova, M.; Rook, L. Dispersal of the Canini (Mammalia, Canidae: Caninae) across Eurasia during the Late Miocene to Early Pleistocene. Quat. Int. 2010, 212, 86–97. [Google Scholar] [CrossRef]
- Wang, X.; Tedford, R.H.; Antón, M. Dogs: Their Fossil Relatives and Evolutionary History; Columbia University Press: New York, NY, USA, 2010. [Google Scholar]
- Puzachenko, A.Y.; Markova, A.K. Diversity dynamics of large- and medium-sized mammals in the Late Pleistocene and the Holocene on the East European Plain: Systems approach. Quat. Int. 2016, 420, 391–401. [Google Scholar] [CrossRef]
- Leonard, J.A.; Vilà, C.; Fox-Dobbs, K.; Koch, P.L.; Wayne, R.K.; van Valkenburgh, B. Megafaunal Extinctions and the Disappearance of a Specialized Wolf Ecomorph. Curr. Biol. 2007, 17, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Skoglund, P.; Ersmark, E.; Palkopoulou, E.; Dalén, L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr. Biol. 2015, 25, 1515–1519. [Google Scholar] [CrossRef] [Green Version]
- Bergström, A.; Stanton, D.W.G.; Taron, U.H.; Frantz, L.; Sinding, M.-H.S.; Ersmark, E.; Pfrengle, S.; Cassatt-Johnstone, M.; Lebrasseur, O.; Girdland-Flink, L.; et al. Grey wolf genomic history reveals a dual ancestry of dogs. Nature 2022, 607, 313–320. [Google Scholar] [CrossRef]
- Vilà, C.; Amorim, I.R.; Leonard, J.A.; Posada, D.; Castroviejo, J.; Petrucci-Fonseca, F.; Crandall, K.A.; Ellegren, H.; Wayne, R.K. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol. Ecol. 1999, 8, 2089–2103. [Google Scholar] [CrossRef] [Green Version]
- Pilot, M.; Branicki, W.; Jędrzejewski, W.; Goszczyński, J.; Jędrzejewska, B.; Dykyy, I.; Shkvyrya, M.; Tsingarska, E. Phylogeographic history of grey wolves in Europe. BMC Evol. Biol. 2010, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.K.; Maldonado, J.E.; Jhala, Y.V.; Fleischer, R.C. Ancient wolf lineages in India. Proc. R. Soc. B Biol. Sci. 2004, 271, 2–5. [Google Scholar] [CrossRef]
- Aggarwal, R.K.; Kivisild, T.; Ramadevi, J.; Singh, L. Mitochondrial DNA coding region sequences support the phylogenetic distinction of two Indian wolf species. J. Zool. Syst. Evol. Res. 2007, 45, 163–172. [Google Scholar] [CrossRef]
- Wayne, R.K.; Lehman, N.; Allard, M.W.; Honeycutt, R.L. Mitochondrial DNA Variability of the Gray Wolf: Genetic Consequences of Population Decline and Habitat Fragmentation. Conserv. Biol. 1992, 6, 559–569. [Google Scholar] [CrossRef]
- Randi, E.; Lucchini, V.; Christensen, M.F.; Mucci, N.; Funk, S.M.; Dolf, G.; Loeschcke, V. Mitochondrial DNA variability in Italian and east European wolves: Detecting the consequences of small population size and hybridization. Conserv. Biol. 2000, 14, 464–473. [Google Scholar] [CrossRef]
- Leonard, J.A.; Vilà, C.; Wayne, R.K. Legacy lost: Genetic variability and population size of extirpated US grey wolves (Canis lupus). Mol. Ecol. 2005, 14, 9–17. [Google Scholar] [CrossRef]
- Weckworth, B.V.; Talbot, S.L.; Cook, J.A. Phylogeography of wolves (Canis lupus) in the Pacific Northwest. J. Mammal. 2010, 91, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, S.A.; Charruau, P.C.; Pollinger, J.P.; Callas, R.; Figura, P.J.; Wayne, R.K. Polyphyletic ancestry of historic gray wolves inhabiting U.S. Pacific states. Conserv. Genet. 2014, 16, 759–764. [Google Scholar] [CrossRef]
- Breitenmoser, U. Large predators in the Alps: The fall and rise of man’s competitors. Biol. Conserv. 1998, 83, 279–289. [Google Scholar] [CrossRef]
- Ripple, W.J.; Estes, J.A.; Beschta, R.L.; Wilmers, C.C.; Ritchie, E.G.; Hebblewhite, M.; Berger, J.; Elmhagen, B.; Letnic, M.; Nelson, M.P.; et al. Status and ecological effects of the world’s largest carnivores. Science 2014, 343, 1241484. [Google Scholar] [CrossRef] [Green Version]
- Boitani, L. Wolf conservation and recovery. In Wolves: Behavior Ecology and Conservation; Mech, L.D., Boitani, L., Eds.; Chicago University Press: Chicago, IL, USA, 2003; pp. 317–340. [Google Scholar]
- Clavero, M.; García-Reyes, A.; Fernández-Gil, A.; Revilla, E.; Fernández, N. Where wolves were: Setting historical baselines for wolf recovery in Spain. Anim. Conserv. 2022. [Google Scholar] [CrossRef]
- Heffelfinger, J.R.; Nowak, R.M.; Paetkau, D. Clarifying historical range to aid recovery of the Mexican wolf. J. Wildl. Manag. 2017, 81, 766–777. [Google Scholar] [CrossRef]
- Hindrikson, M.; Remm, J.; Pilot, M.; Godinho, R.; Stronen, A.V.; Baltrūnaité, L.; Czarnomska, S.D.; Leonard, J.A.; Randi, E.; Nowak, C.; et al. Wolf population genetics in Europe: A systematic review, meta-analysis and suggestions for conservation and management. Biol. Rev. 2017, 92, 1601–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinding, M.-H.S.; Gopalakrishan, S.; Vieira, F.G.; Samaniego Castruita, J.A.; Raundrup, K.; Heide Jørgensen, M.P.; Meldgaard, M.; Petersen, B.; Sicheritz-Ponten, T.; Mikkelsen, J.B.; et al. Population genomics of grey wolves and wolf-like canids in North America. PLoS Genet. 2018, 14, e1007745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taron, U.H.; Salado, I.; Escobar-Rodríguez, M.; Westbury, M.V.; Butschkau, S.; Paijmans, J.L.A.; von Holdt, B.M.; Hofreiter, M.; Leonard, J.A. A sliver of the past: The decimation of the genetic diversity of the Mexican wolf. Mol. Ecol. 2021, 30, 6340–6354. [Google Scholar] [CrossRef] [PubMed]
- Jansson, E.; Harmoinen, J.; Ruokonen, M.; Aspi, J. Living on the edge: Reconstructing the genetic history of the Finnish wolf population. BMC Evol. Biol. 2014, 14, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufresnes, C.; Miquel, C.; Remollino, N.; Biollaz, F.; Salamin, N.; Taberlet, P.; Fumagalli, L. Howling from the past: Historical phylogeography and diversity losses in European grey wolves. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181148. [Google Scholar] [CrossRef] [Green Version]
- Dufresnes, C.; Miquel, C.; Taberlet, P.; Fumagalli, L. Last but not beast: The fall of the Alpine wolves told by historical DNA. Mammal Res. 2019, 64, 595–600. [Google Scholar] [CrossRef]
- Valverde, J.A. El lobo español. Montes 1971, 159, 228–241. [Google Scholar]
- del Grande Brío, R. El Lobo Ibérico. Biología y Mitología; Hermann Blume: Madrid, Spain, 1984. [Google Scholar]
- Blanco, J.C.; Cuesta, L.; Reig, S. El lobo en España: Una visión global. In El lobo (Canis lupus) en España. Situación, Problemática y Apuntes Sobre Su Ecología; Blanco, J.C., Cuesta, L., Reig, S., Eds.; Ministerio de Agriculura, Pesca y Alimentación (ICONA): Madrid, Spain, 1990; pp. 69–82. [Google Scholar]
- Gómez-Sánchez, D.; Olalde, I.; Sastre, N.; Enseñat, C.; Carrasco, R.; Marques-Bonet, T.; Lalueza-Fox, C.; Leonard, J.A.; Vilà, C.; Ramírez, O. On the path to extinction: Inbreeding and admixture in a declining gray wolf population. Mol. Ecol. 2018, 27, 3599–3612. [Google Scholar] [CrossRef]
- Chapron, G.; Kaczensky, P.; Linnell, J.D.C.; von Arx, M.; Huber, D.; Andrén, H.; López-bao, J.V.; Adamec, M. Recovery of large carnivores in Europe’ s modern human-dominated landscapes. Science 2014, 346, 1517–1519. [Google Scholar] [CrossRef] [Green Version]
- MITECO. Censo 2012–2014 de Lobo Ibérico (Canis lupus Linnaeus 1758) en España; MITECO: Madrid, Spain, 2016. [Google Scholar]
- Bencatel, J.; Sabino-Marques, H.; Álvares, F.; Moura, A.E.; Barbosa, A.M. Atlas de Mamíferos de Portugal, 2nd ed.; Universidad de Évora: Évora, Portugal, 2019. [Google Scholar]
- Gutiérrez Alba, V. El Lobo Ibérico en Andalucía: Historia, Mitología, Relaciones con el Hombre; Junta de Andalucía; Fundación Gypaetus: Jaén, Spain, 2006. [Google Scholar]
- de Andalucía, J. Memoria Técnica Justificativa para la Catalogación del Lobo Ibérico en Andalucía; Consejería de Medio Ambiente y Ordenación del Territorio; Junta de Andalucía: Seville, Spain, 2015. [Google Scholar]
- López-Bao, J.V.; Fleurke, F.; Chapron, G.; Trouwborst, A. Legal obligations regarding populations on the verge of extinction in Europe: Conservation, Restoration, Recolonization, Reintroduction. Biol. Conserv. 2018, 227, 319–325. [Google Scholar] [CrossRef]
- Ciucani, M.M.; Palumbo, D.; Galaverni, M.; Serventi, P.; Fabbri, E.; Ravegnini, G.; Angelini, S.; Maini, E.; Persico, D.; Caniglia, R.; et al. Old wild wolves: Ancient DNA survey unveils population dynamics in Late Pleistocene and Holocene Italian remains. PeerJ 2019, 2019, e6424. [Google Scholar] [CrossRef]
- Knapp, M.; Hofreiter, M. Next Generation Sequencing of Ancient DNA: Requirements, Strategies and Perspectives. Genes 2010, 1, 227–243. [Google Scholar] [CrossRef] [Green Version]
- Losey, R.J.; Bazaliiskii, V.I.; Garvie-Lok, S.; Germonpré, M.; Leonard, J.A.; Allen, A.L.; Anne Katzenberg, M.; Sablin, M.V. Canids as persons: Early Neolithic dog and wolf burials, Cis-Baikal, Siberia. J. Anthropol. Archaeol. 2011, 30, 174–189. [Google Scholar] [CrossRef] [Green Version]
- Byrd, B.F.; Cornellas, A.; Eerkens, J.W.; Rosenthal, J.S.; Carpenter, T.R.; Leventhal, A.; Leonard, J.A. The role of canids in ritual and domestic contexts: New ancient DNA insights from complex hunter–gatherer sites in prehistoric Central California. J. Archaeol. Sci. 2013, 40, 2176–2189. [Google Scholar] [CrossRef] [Green Version]
- Hagelberg, E.; Hofreiter, M.; Keyser, C. Ancient DNA: The first three decades. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20130371. [Google Scholar] [CrossRef] [Green Version]
- Björnerfeldt, S.; Webster, M.T.; Vilà, C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006, 16, 990–994. [Google Scholar] [CrossRef] [Green Version]
- Koblmüller, S.; Vilà, C.; Lorente-Galdos, B.; Dabad, M.; Ramirez, O.; Marques-Bonet, T.; Wayne, R.K.; Leonard, J.A. Whole mitochondrial genomes illuminate ancient intercontinental dispersals of grey wolves (Canis lupus). J. Biogeogr. 2016, 43, 1728–1738. [Google Scholar] [CrossRef]
- Loog, L.; Thalmann, O.; Sinding, M.S.; Schuenemann, V.J.; Perri, A.; Germonpré, M.; Bocherens, H.; Witt, K.E.; Samaniego Castruita, J.A.; Velasco, M.S.; et al. Ancient DNA suggests modern wolves trace their origin to a Late Pleistocene expansion from Beringia. Mol. Ecol. 2020, 29, 1596–1610. [Google Scholar] [CrossRef] [Green Version]
- Thalmann, O.; Shapiro, B.; Cui, P.; Schuenemann, V.J.; Sawyer, S.K.; Greenfield, D.L.; Germonpré, M.B.; Sablin, M.V.; López-Giráldez, F.; Domingo-Roura, X.; et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 2013, 342, 871–874. [Google Scholar] [CrossRef] [Green Version]
- Camacho-Sanchez, M.; Burraco, P.; Gomez-Mestre, I.; Leonard, J.A. Preservation of RNA and DNA from mammal samples under field conditions. Mol. Ecol. Resour. 2013, 13, 663–673. [Google Scholar] [CrossRef] [Green Version]
- Blaschikoff, L.; Daza-Perea, A.; Requicha, J.; Detry, C.; Rasteiro, R.; Guimarães, S.; Ureña, I.; Serra, O.; Schmidt, R.; Valera, A.; et al. A multidisciplinary study of Iberian Chalcolithic dogs. J. Archaeol. Sci. Rep. 2022, 42, 103338. [Google Scholar] [CrossRef]
- Koepfli, K.P.; Pollinger, J.; Godinho, R.; Robinson, J.; Lea, A.; Hendricks, S.; Schweizer, R.M.; Thalmann, O.; Silva, P.; Fan, Z.; et al. Genome-wide evidence reveals that African and Eurasian golden jackals are distinct species. Curr. Biol. 2015, 25, 2158–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Silva, P.; Gronau, I.; Wang, S.; Armero, A.S.; Schweizer, R.M.; Ramirez, O.; Pollinger, J.; Galaverni, M.; Del-Vecchyo, D.O.; et al. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 2016, 26, 163–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SNPFC. Lobo (Canis lupus). Mapa Cinegético Nacional. Avance Informativo; Servicio Nacional de Pesca Fluvial y Caza: Madrid, Spain, 1968. [Google Scholar]
- Petrucci-Fonseca, F. O lobo (Canis lupus signatus Cabrera, 1907) em Portugal. Problemática da Sua Conservação. Ph.D. Dissertation, Faculdade de Ciências da Universidade de Lisboa, Lisbon, Portugal, 1990. [Google Scholar]
- Ford, B.M.; Cornellas, A.; Leonard, J.A.; Weir, R.D.; Russello, M.A. Spatiotemporal analyses suggest the role of glacial history and the ice-free corridor in shaping American badger population genetic variation. Ecol. Evol. 2020, 10, 8345–8357. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Lindqvist, C. Technical advances and challenges in genome-scale analysis of ancient DNA. In Paleogenomics: Genome-Scale Analysis of Ancient DNA; Lindqvist, C., Rajora, O.P., Eds.; Springer: Cham, Switzerland, 2019; pp. 3–30. [Google Scholar]
- Carøe, C.; Gopalakrishnan, S.; Lasse Vinner, L.; Mak, S.S.T.; Sinding, M.H.S.; Samaniego, J.A.; Wales, N.; Sicheritz-Pontén, T.; Gilber, M.T.P. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 2018, 9, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Mak, S.S.T.; Gopalakrishnan, S.; Carøe, C.; Geng, C.; Liu, S.; Sinding, M.H.S.; Kuderna, L.F.K.; Zhang, W.; Fu, S.; Vieira, F.G.; et al. Comparative performance of the BGISEQ-500 vs. Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience 2017, 6, gix049. [Google Scholar] [CrossRef] [Green Version]
- Gansauge, M.T.; Meyer, M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 2013, 8, 737–748. [Google Scholar] [CrossRef]
- Paijmans, J.L.A.; Baleka, S.; Henneberger, K.; Taron, U.H.; Trinks, A.; Westbury, M.V.; Barlow, A. Sequencing Single-Stranded Libraries on the Illumina NextSeq 500 Platform. arXiv 2017. [Google Scholar] [CrossRef]
- Neiman, M.; Sundling, S.; Grönberg, H.; Hall, P.; Czene, K.; Lindberg, J.; Klevebring, D. Library Preparation and Multiplex Capture for Massive Parallel Sequencing Applications Made Efficient and Easy. PLoS ONE 2012, 7, e48616. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 October 2021).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Li, G.; Davis, B.W.; Raudsepp, T.; Wilkerson, A.J.P.; Mason, V.C.; Ferguson-Smith, M.; O’Brien, P.C.; Waters, P.D.; Murphy, W.J. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 2013, 23, 1486–1495. [Google Scholar] [CrossRef] [Green Version]
- den Tex, R.J.; Maldonado, J.E.; Thorington, R.; Leonard, J.A. Nuclear copies of mitochondrial genes: Another problem for ancient DNA. Genetica 2010, 138, 979–984. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Brown, S.D.J.; Collins, R.A.; Boyer, S.; Lefort, M.C.; Malumbres-Olarte, J.; Vink, C.J.; Cruickshank, R.H. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 2012, 12, 562–565. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http//www.R-project.org/ (accessed on 1 November 2022).
- Wickham, H. GGPLOT2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 9783319242774. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Alexander, A.; Steel, D.; Hoekzema, K.; Mesnick, S.L.; Engelhaupt, D.; Kerr, I.; Payne, R.; Baker, C.S. What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol. Ecol. 2016, 25, 2754–2772. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J. FigTree Version 1.4.4. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, Scotland, 2018. [Google Scholar]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ramirez, O.; Altet, L.; Enseñat, C.; Vilà, C.; Sanchez, A.; Ruiz, A. Genetic assessment of the Iberian wolf Canis lupus signatus captive breeding program. Conserv. Genet. 2006, 7, 861–878. [Google Scholar] [CrossRef] [Green Version]
- Sastre, N.; Vilà, C.; Salinas, M.; Bologov, V.V.; Urios, V.; Sánchez, A.; Francino, O.; Ramírez, O. Signatures of demographic bottlenecks in European wolf populations. Conserv. Genet. 2011, 12, 701–712. [Google Scholar] [CrossRef]
- Matsumura, S.; Inoshima, Y.; Ishiguro, N. Reconstructing the colonization history of lost wolf lineages by the analysis of the mitochondrial genome. Mol. Phylogenet. Evol. 2014, 80, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Pilot, M.; Jedrzejewski, W.; Branicki, W.; Sidorovich, V.E.; Jedrzejewska, B.; Stachura, K.; Funk, S.M. Ecological factors influence population genetic structure of European grey wolves. Mol. Ecol. 2006, 15, 4533–4553. [Google Scholar] [CrossRef] [PubMed]
- Musiani, M.; Leonard, J.A.; Cluff, H.D.; Gates, C.C.; Mariani, S.; Paquet, P.C.; Vilà, C.; Wayne, R.K. Differentiation of tundra/taiga and boreal coniferous forest wolves: Genetics, coat colour and association with migratory caribou. Mol. Ecol. 2007, 16, 4149–4170. [Google Scholar] [CrossRef]
- Muñoz-Fuentes, V.; Darimont, C.T.; Wayne, R.K.; Paquet, P.C.; Leonard, J.A. Ecological factors drive differentiation in wolves from British Columbia. J. Biogeogr. 2009, 36, 1516–1531. [Google Scholar] [CrossRef] [Green Version]
- Ersmark, E.; Klütsch, C.F.C.; Chan, Y.L.; Sinding, M.H.S.; Fain, S.R.; Illarionova, N.A.; Oskarsson, M.; Uhlén, M.; Zhang, Y.P.; Dalén, L.; et al. From the past to the present: Wolf phylogeography and demographic history based on the mitochondrial control region. Front. Ecol. Evol. 2016, 4, 134. [Google Scholar] [CrossRef] [Green Version]
- Leonard, J.A.; Wayne, R.K.; Wheeler, J.; Valadez, R.; Guillén, S.; Vilà, C. Ancient DNA evidence for Old World origin of New World dogs. Science 2002, 298, 1613–1616. [Google Scholar] [CrossRef] [Green Version]
- KoblmÜller, S.; Nord, M.; Wayne, R.K.; Leonard, J.A. Origin and status of the Great Lakes wolf. Mol. Ecol. 2009, 18, 2313–2326. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Fuentes, V.; Darimont, C.T.; Paquet, P.C.; Leonard, J.A. The genetic legacy of extirpation and re-colonization in Vancouver Island wolves. Conserv. Genet. 2010, 11, 547–556. [Google Scholar] [CrossRef]
- Losey, R.J.; Garvie-Lok, S.; Leonard, J.A.; Katzenberg, M.A.; Germonpré, M.; Nomokonova, T.; Sablin, M.V.; Goriunova, O.I.; Berdnikova, N.E.; Savel’ev, N.A. Burying Dogs in Ancient Cis-Baikal, Siberia: Temporal Trends and Relationships with Human Diet and Subsistence Practices. PLoS ONE 2013, 8, e63740. [Google Scholar] [CrossRef] [Green Version]
- Keis, M.; Remm, J.; Ho, S.Y.W.; Davison, J.; Tammeleht, E.; Tumanov, I.L.; Saveljev, A.P.; Männil, P.; Kojola, I.; Abramov, A.V.; et al. Complete mitochondrial genomes and a novel spatial genetic method reveal cryptic phylogeographical structure and migration patterns among brown bears in north-western Eurasia. J. Biogeogr. 2013, 40, 915–927. [Google Scholar] [CrossRef]
- Rodríguez, R.; Ramírez, O.; Valdiosera, C.E.; García, N.; Alda, F.; Madurell-Malapeira, J.; Marmi, J.; Doadrio, I.; Willerslev, E.; Götherström, A.; et al. 50,000 years of genetic uniformity in the critically endangered Iberian lynx. Mol. Ecol. 2011, 20, 3785–3795. [Google Scholar] [CrossRef] [Green Version]
- Casas-Marce, M.; Marmesat, E.; Soriano, L.; Martínez-Cruz, B.; Lucena-Perez, M.; Nocete, F.; Rodríguez-Hidalgo, A.; Canals, A.; Nadal, J.; Detry, C.; et al. Spatiotemporal Dynamics of Genetic Variation in the Iberian lynx along Its Path to Extinction Reconstructed with Ancient DNA. Mol. Biol. Evol. 2017, 34, 2893–2907. [Google Scholar] [CrossRef] [Green Version]
- Valdiosera, C.E.; García-Garitagoitia, J.L.; Garcia, N.; Doadrio, I.; Thomas, M.G.; Hänni, C.; Arsuaga, J.L.; Barnes, I.; Hofreiter, M.; Orlando, L.; et al. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos). Proc. Natl. Acad. Sci. USA 2008, 105, 5123–5128. [Google Scholar] [CrossRef] [Green Version]
- Blanco, J.C.; Rodriguez, A.; Cuesta, L.; Reig, S.; del Olmo, J.C. El Lobo En Sierra Morena. In El lobo (Canis lupus) en España. Situación, Problemática y Apuntes Sobre Su Ecología; Blanco, J.C., Cuesta, L., Reig, S., Eds.; Ministerio de Agriculura, Pesca y Alimentación (ICONA): Madrid, Spain, 1990; pp. 61–68. [Google Scholar]
- Quevedo, M.; Echegaray, J.; Fernández-Gil, A.; Leonard, J.A.; Naves, J.; Ordiz, A.; Revilla, E.; Vilà, C. Lethal management may hinder population recovery in Iberian wolves. Biodivers. Conserv. 2019, 28, 415–432. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salado, I.; Preick, M.; Lupiáñez-Corpas, N.; Fernández-Gil, A.; Vilà, C.; Hofreiter, M.; Leonard, J.A. Loss of Mitochondrial Genetic Diversity despite Population Growth: The Legacy of Past Wolf Population Declines. Genes 2023, 14, 75. https://doi.org/10.3390/genes14010075
Salado I, Preick M, Lupiáñez-Corpas N, Fernández-Gil A, Vilà C, Hofreiter M, Leonard JA. Loss of Mitochondrial Genetic Diversity despite Population Growth: The Legacy of Past Wolf Population Declines. Genes. 2023; 14(1):75. https://doi.org/10.3390/genes14010075
Chicago/Turabian StyleSalado, Isabel, Michaela Preick, Natividad Lupiáñez-Corpas, Alberto Fernández-Gil, Carles Vilà, Michael Hofreiter, and Jennifer A. Leonard. 2023. "Loss of Mitochondrial Genetic Diversity despite Population Growth: The Legacy of Past Wolf Population Declines" Genes 14, no. 1: 75. https://doi.org/10.3390/genes14010075