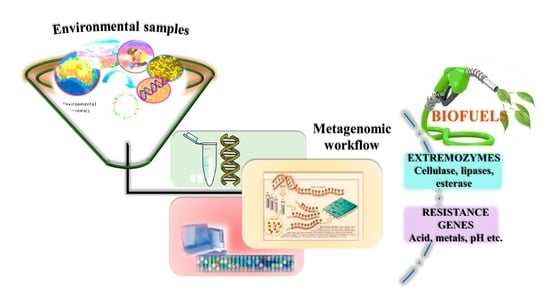
Unravelling Metagenomics Approach for Microbial Biofuel Production
Abstract
:1. Introduction
2. Microbial Biofuel—An Overview
3. Introduction of Metagenomics
4. Metagenomics—An Advanced Tool in Biofuel Sector
5. Metagenomics and Enzyme Engineering Assisted Upscaling of Biofuel Production
6. Metagenomic Implications on Biotechnology
7. Future Perspectives and Challenges
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, A.; Sarkar, O.; Rova, U.; Christakopoulos, P.; Matsakas, L. Valorization of volatile fatty acids derived from low-cost organic waste for lipogenesis in oleaginous microorganisms-A review. Bioresour. Technol. 2021, 321, 124457. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Zhang, X.; Huang, H. Application of metagenomic techniques in mining enzymes from microbial communities for biofuel synthesis. Biotechnol. Adv. 2012, 30, 920–929. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Joint Statement between the European Commission and the United States on European Energy Security; European Commission: Brussels, Belgium, 2022. [Google Scholar]
- Patel, A.; Arora, N.; Sartaj, K.; Pruthi, V.; Pruthi, P.A. Sustainable biodiesel production from oleaginous yeasts utilizing hydrolysates of various non-edible lignocellulosic biomasses. Renew. Sustain. Energy Rev. 2016, 62, 836–855. [Google Scholar] [CrossRef]
- Madhavan, A.; Sindhu, R. Metagenome Analysis: A Powerful Tool for Enzyme Bioprospecting. Appl. Biochem. Biotechnol. 2017, 183, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022, 10, 76. [Google Scholar] [CrossRef]
- Biddle, J.F.; Fitz-gibbon, S.; Schuster, S.C.; Brenchley, J.E.; House, C.H. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. USA 2008, 105, 10583–10588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.; Nelson, E.; Tilman, D.; Polasky, S.; Tiffany, D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. USA 2006, 103, 11206–11210. [Google Scholar] [CrossRef] [Green Version]
- Mohr, A.; Raman, S. Lessons from first generation biofuels and implications for the sustainability appraisal of second generation biofuels. Energy Policy 2015, 63, 281–310. [Google Scholar] [CrossRef]
- Ramos, J.L.; Pakuts, B.; Godoy, P.; García-Franco, A.; Duque, E. Addressing the energy crisis: Using microbes to make biofuels. Microb. Biotechnol. 2022, 15, 1026–1030. [Google Scholar] [CrossRef]
- Subhash, G.V.; Mohan, S.V. Biodiesel production from isolated oleaginous fungi Aspergillus sp. using corncob waste liquor as a substrate. Bioresour. Technol. 2011, 102, 9286–9290. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Guan, M.; Tang, H.; Wang, Z.; Lin, L.; Pang, H. Production of butanol from lignocellulosic biomass: Recent advances, challenges, and prospects. RSC Adv. 2022, 12, 18848–18863. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Al Makishah, N.H.; Wen, Z.; Gupta, G.; Pandit, S.; Prasad, R. Recent Developments in Lignocellulosic Biofuels, a Renewable Source of Bioenergy. Fermentation 2022, 8, 161. [Google Scholar] [CrossRef]
- Datta, S.; Rajnish, K.N.; Samuel, M.S.; Pugazlendhi, A.; Selvarajan, E. Metagenomic applications in microbial diversity, bioremediation, pollution monitoring, enzyme and drug discovery. Environ. Chem. Lett. 2020, 18, 1229–1241. [Google Scholar] [CrossRef]
- Alves, L.D.F.; Westmann, C.A.; Lovate, G.L.; Marcelino, G.; De Siqueira, V.; Borelli, T.C.; Guazzaroni, M. Metagenomic Approaches for Understanding New Concepts in Microbial Science. Int. J. Genom. 2018, 2018, 2312987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Hart, D.J.; An, Y.; Hart, D.J. Functional Metagenomic Technologies for the Discovery of Novel Enzymes for Biomass Degradation and Biofuel Production. Bioenergy Res. 2019, 12, 457–470. [Google Scholar] [CrossRef]
- Shah, R.K.; Patel, A.K.; Davla, D.M.; Parikh, I.K. Molecular cloning, heterologous expression, and functional characterization of a cellulolytic enzyme (Cel PRII) from buffalo rumen metagenome. 3 Biotech 2017, 7, 257. [Google Scholar] [CrossRef]
- Yang, C.; Xia, Y.; Qu, H.; Li, A.D.; Liu, R.; Wang, Y.; Zhang, T. Discovery of new cellulases from the metagenome by a metagenomics—Guided strategy. Biotechnol. Biofuels 2016, 9, 138. [Google Scholar] [CrossRef] [Green Version]
- Maruthamuthu, M.; Jiménez, D.J.; Stevens, P.; Van, E.J.D. A multi-substrate approach for functional metagenomics-based screening for (hemi) cellulases in two wheat straw-degrading microbial consortia unveils novel thermoalkaliphilic enzymes. BMC Genom. 2016, 17, 86. [Google Scholar] [CrossRef] [Green Version]
- Hung, K.S.; Liu, S.M.; Tzou, W.S.; Lin, F.P.; Pan, C.L.; Fang, T.Y.; Sun, K.H.; Tang, S.J. Characterization of a novel GH10 thermostable, halophilic xylanase from the marine bacterium Thermoanaerobacterium saccharolyticum NTOU1. Process Biochem. 2011, 46, 1257–1263. [Google Scholar] [CrossRef]
- Findley, S.D.; Mormile, M.R.; Sommer-Hurley, A.; Zhang, X.C.; Tipton, P.; Arnett, K.; Porter, J.H.; Kerley, M.; Stacey, G. Activity-Based Metagenomic Screening and Biochemical Characterization of Bovine Ruminal Protozoan Glycoside Hydrolases. Appl. Environ. Microbiol. 2011, 77, 8106–8113. [Google Scholar] [CrossRef]
- Ausec, L.; Berini, F.; Casciello, C.; Cretoiu, M.S.; van Elsas, J.D.; Marinelli, F.; Mandic-Mulec, I. The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and thermo-tolerance. Appl. Microbiol. Biotechnol. 2017, 101, 6261–6276. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Hu, Y.; Xie, F.; Guo, H.; Ashforth, E.J.; Polyak, S.W.; Zhu, B.; Zhang, L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 2011, 90, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Ju, F.; Fang, H.H.P.; Zhang, T. Mining of Novel Thermo-Stable Cellulolytic Genes from a Thermophilic Cellulose-Degrading Consortium by Metagenomics. PLoS ONE 2013, 8, 53779. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, M.; Martínez-Abarca, F.; Golyshin, P.N. Mining genomes and “metagenomes” for novel catalysts. Curr. Opin. Biotechnol. 2005, 16, 588–593. [Google Scholar] [CrossRef]
- Ono, A.; Miyazaki, R.; Sota, M.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Isolation and characterization of naphthalene-catabolic genes and plasmids from oil-contaminated soil by using two cultivation-independent approaches. Appl. Microbiol. Biotechnol. 2007, 74, 501–510. [Google Scholar] [CrossRef]
- Nagayama, H.; Sugawara, T.; Endo, R.; Ono, A.; Kato, H.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Isolation of oxygenase genes for indigo-forming activity from an artificially polluted soil metagenome by functional screening using Pseudomonas putida strains as hosts. Appl. Microbiol. Biotechnol. 2015, 99, 4453–4470. [Google Scholar] [CrossRef]
- Fang, Z.; Li, T.; Wang, Q.; Zhang, X.; Peng, H.; Fang, W.; Hong, Y.; Ge, H.; Xiao, Y. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl. Microbiol. Biotechnol. 2011, 89, 1103–1110. [Google Scholar] [CrossRef]
- Silva, C.C.; Hayden, H.; Sawbridge, T.; Mele, P.; De Paula, S.O.; Silva, L.C.F.; Vidigal, P.M.P.; Vicentini, R.; Sousa, M.P.; Torres, A.P.R.; et al. Identification of Genes and Pathways Related to Phenol Degradation in Metagenomic Libraries from Petroleum Refinery Wastewater. PLoS ONE 2013, 8, e61811. [Google Scholar] [CrossRef] [Green Version]
- Thies, S.; Rausch, S.C.; Kovacic, F.; Schmidt-Thaler, A.; Wilhelm, S.; Rosenau, F.; Daniel, R.; Streit, W.; Pietruszka, J.; Jaeger, K.E. Metagenomic discovery of novel enzymes and biosurfactants in a slaughterhouse biofilm microbial community. Sci. Rep. 2016, 6, 27035. [Google Scholar] [CrossRef] [Green Version]
- Schröder, C.; Elleuche, S.; Blank, S.; Antranikian, G. Characterization of a heat-active archaeal β-glucosidase from a hydrothermal spring metagenome. Enzym. Microb. Technol. 2014, 57, 48–54. [Google Scholar] [CrossRef]
- Zarafeta, D.; Kissas, D.; Sayer, C.; Gudbergsdottir, S.R.; Ladoukakis, E.; Isupov, M.N.; Chatziioannou, A.; Peng, X.; Littlechild, J.A.; Skretas, G.; et al. Discovery and Characterization of a Thermostable and Highly Halotolerant GH5 Cellulase from an Icelandic Hot Spring Isolate. PLoS ONE 2016, 11, e0146454. [Google Scholar] [CrossRef] [PubMed]
- Bilal, T.; Malik, B.; Rehman, K. Metagenomic analysis of uncultured microorganisms and their enzymatic attributes. J. Microbiol. Methods 2018, 155, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Kanafusa-shinkai, S.; Wakayama, J.; Tsukamoto, K.; Hayashi, N.; Miyazaki, Y.; Ohmori, H.; Tajima, K.; Yokoyama, H. Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium Caldicellulosiruptor bescii. J. Biosci. Bioeng. 2013, 115, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Aylward, F.O.; Burnum, K.E.; Scott, J.J.; Suen, G.; Tringe, S.G.; Adams, S.M.; Barry, K.W.; Nicora, C.D.; Piehowski, P.D.; Purvine, S.O.; et al. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J. 2012, 6, 1688–1701. [Google Scholar] [CrossRef] [Green Version]
- Duan, C.; Xian, L.; Zhao, G.; Feng, Y.; Pang, H.; Bai, X.; Tang, J.; Ma, Q.; Feng, J. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J. Appl. Microbiol. 2009, 107, 245–256. [Google Scholar] [CrossRef]
- Fang, W.; Liu, J.; Hong, Y.; Peng, H.; Zhang, X.; Sun, B.; Xiao, Y. Cloning and characterization of a β-glucosidase from marine microbial metagenome with excellent glucose tolerance. J. Microbiol. Biotechnol. 2010, 20, 1351–1358. [Google Scholar] [CrossRef]
- Jiang, C.; Li, S.; Luo, F.; Jin, K.; Wang, Q.; Hao, Z.; Wu, L.; Zhao, G.; Ma, G.; Shen, P.; et al. Biochemical characterization of two novel β-glucosidase genes by metagenome expression cloning. Bioresour. Technol. 2011, 102, 3272–3278. [Google Scholar] [CrossRef]
- Beloqui, A.; Nechitaylo, T.Y.; Lo, N.; Polaina, J.; Strittmatter, A.W.; Reva, O.; Waliczek, A.; Yakimov, M.M.; Golyshina, O.V.; Ferrer, M.; et al. Diversity of Glycosyl Hydrolases from Cellulose-Depleting Communities Enriched from Casts of Two Earthworm Species. Appl. Environ. Microbiol. 2010, 76, 5934–5946. [Google Scholar] [CrossRef] [Green Version]
- Brennan, Y.; Callen, W.N.; Christoffersen, L.; Dupree, P.; Goubet, F.; Healey, S.; Herna, M.; Keller, M.; Li, K.; Palackal, N.; et al. Unusual Microbial Xylanases from Insect Guts. Appl. Environ. Microbiol. 2004, 70, 3609–3617. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.C.; Kibblewhite-accinelli, R.E.; Wagschal, K.; Robertson, G.H.; Wong, D.W.S. Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extreamophiles 2006, 10, 295–300. [Google Scholar] [CrossRef]
- Liu, N.; Yan, X.; Zhang, M.; Xie, L.; Wang, Q.; Huang, Y.; Zhou, X.; Wang, S.; Zhou, Z. Microbiome of fungus-growing termites: A new reservoir for lignocellulase genes. Appl. Environ. Microbiol. 2011, 77, 48–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, M.; Li, G.; Liang, W.Q.; Liu, Y.H. Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl. Microbiol. Biotechnol. 2010, 87, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Beloqui, A.; Pita, M.; Polaina, J.; Martı, A.; Golyshina, O.V.; Zuma, M.; Yakimov, M.M.; Garcı, H.; Alcalde, M.; Ferna, M.; et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen biochemical properties, structural analysis, and phylogenetic relationship. J. Biol. Chem. 2006, 281, 22933–22942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Oh, K.; Lee, M.; Kang, C.; Oh, T.; Yoon, J. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol. 2009, 75, 257–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.H.; Kim, J.; Kim, Y.J.; Kim, H.; Lee, H.S.; Kang, S.G.; Kim, S.; Lee, J. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009, 81, 865–874. [Google Scholar] [CrossRef]
- Elend, C.; Schmeisser, C.; Hoebenreich, H.; Steele, H.L.; Streit, W.R. Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J. Biotechnol. 2007, 130, 370–377. [Google Scholar] [CrossRef]
- Bunterngsook, B.; Kanokratana, P.; Tanapongpipat, S.; Uengwetwanit, T.; Rachdawong, S.; Vichitsoonthonkul, T.; Anapongpipat, S.T.; Engwetwanit, T.U.; Achdawong, S.R. Identification and characterization of lipolytic enzymes from a peat-swamp forest soil metagenome. Biosci. Biotechnol. Biochem. 2010, 74, 1848–1854. [Google Scholar] [CrossRef] [Green Version]
- Rhee, J.K.; Ahn, D.G.; Kim, Y.G.; Oh, J.W. New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl. Environ. Microbiol. 2005, 71, 817–825. [Google Scholar] [CrossRef] [Green Version]
- Tirawongsaroj, P.; Sriprang, R.; Harnpicharnchai, P.; Thongaram, T.; Champreda, V.; Tanapongpipat, S. Novel thermophilic and thermostable lipolytic enzymes from a thailand hot spring metagenomic library. J. Biotechnol. 2008, 133, 42–49. [Google Scholar] [CrossRef]
- Yu, E.Y.; Kwon, M.; Lee, M. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl. Microbiol. Biotechnol. 2011, 90, 573–581. [Google Scholar] [CrossRef]
- Wexler, M.; Bond, P.L.; Richardson, D.J.; Johnston, A.W.B. A wide host-range metagenomic library from a waste water treatment plant yields a novel alcohol/aldehyde dehydrogenase. Environ. Microbiol. 2005, 7, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Knietsch, A.; Waschkowitz, T.; Bowien, S.; Henne, A.; Daniel, R. Construction and Screening of Metagenomic Libraries Derived from Enrichment Cultures: Generation of a Gene Bank for Genes Conferring Alcohol Oxidoreductase Activity on Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 1408–1416. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Mishra, S.D. Recent advances in biofuel production through metabolic engineering. Bioresour. Technol. 2022, 352, 127037. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Huang, S.; Liu, T.; Geng, A. Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb. Cell Fact. 2015, 14, 70. [Google Scholar] [CrossRef] [Green Version]
- Parachin, N.S.; Gorwa-Grauslund, M.F. Isolation of xylose isomerases by sequence-and function-based screening from a soil metagenomic library. Biotechnol. Biofuels 2011, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Shen, Y.; Jiao, C.; Ge, R.; Zhang, X.; Bao, X. Characterization and evolution of xylose isomerase screened from the bovine rumen metagenome in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2016, 121, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Katahira, S.; Muramoto, N.; Moriya, S.; Nagura, R.; Tada, N.; Yasutani, N.; Ohkuma, M.; Onishi, T.; Tokuhiro, K. Screening and evolution of a novel protist xylose isomerase from the termite Reticulitermes speratus for efficient xylose fermentation in Saccharomyces cerevisiae. Biotechnol. Biofuels 2017, 10, 203. [Google Scholar] [CrossRef] [Green Version]
- Rajabi, M.; Nourisanami, F.; Ghadikolaei, K.K.; Changizian, M.; Noghabi, K.A.; Zahiri, H.S. Metagenomic psychrohalophilic xylanase from camel rumen investigated for bioethanol production from wheat bran using Bacillus subtilis AP. Sci. Rep. 2022, 12, 8152. [Google Scholar] [CrossRef]
- Ariaeenejad, S.; Sheykh Abdollahzadeh Mamaghani, A.; Maleki, M.; Kavousi, K.; Foroozandeh Shahraki, M.; Hosseini Salekdeh, G. A novel high performance in-silico screened metagenome-derived alkali-thermostable endo-β-1,4-glucanase for lignocellulosic biomass hydrolysis in the harsh conditions. BMC Biotechnol. 2020, 20, 56. [Google Scholar] [CrossRef]
- Patel, M.; Patel, H.M.; Dave, S. Determination of bioethanol production potential from lignocellulosic biomass using novel Cel-5m isolated from cow rumen metagenome. Int. J. Biol. Macromol. 2020, 153, 1099–1106. [Google Scholar] [CrossRef]
- Escuder-Rodríguez, J.J.; González-Suarez, M.; deCastro, M.E.; Saavedra-Bouza, A.; Becerra, M.; González-Siso, M.I. Characterization of a novel thermophilic metagenomic GH5 endoglucanase heterologously expressed in Escherichia coli and Saccharomyces cerevisiae. Biotechnol. Biofuels Bioprod. 2022, 15, 76. [Google Scholar] [CrossRef] [PubMed]
- Giannakopoulou, A.; Patila, M.; Spyrou, K.; Chalmpes, N.; Zarafeta, D.; Skretas, G.; Gournis, D.; Stamatis, H. Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions. Catalysts 2019, 9, 995. [Google Scholar] [CrossRef] [Green Version]
- Geng, A.; Zou, G.; Yan, X.; Wang, Q.; Zhang, J.; Liu, F.; Zhu, B.; Zhou, Z. Expression and characterization of a novel metagenome-derived cellulase Exo2b and its application to improve cellulase activity in Trichoderma reesei. Appl. Microbiol. Biotechnol. 2012, 96, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Madalozzo, A.D.; Martini, V.P.; Kuniyoshi, K.K.; De Souza, E.M.; Pedrosa, F.O.; Glogauer, A.; Zanin, G.M.; Mitchell, D.A.; Krieger, N. Immobilization of LipC12, a new lipase obtained by metagenomics, and its application in the synthesis of biodiesel esters. J. Mol. Catal. B Enzym. 2015, 116, 45–51. [Google Scholar] [CrossRef]
- Yan, W.; Li, F.; Wang, L.; Zhu, Y.; Dong, Z.; Bai, L. Discovery and characterizaton of a novel lipase with transesterification activity from hot spring metagenomic library. Biotechnol. Rep. 2017, 14, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Shestakov, S.V. Impact of Metagenomics on Biotechnological Development. Appl. Biochem. Microbiol. 2012, 48, 705–715. [Google Scholar] [CrossRef]
- Li, L.; Mccorkle, S.R.; Monchy, S.; Taghavi, S.; Lelie, D.V.D. Bioprospecting metagenomes: Glycosyl hydrolases for converting biomass. Biotechnol. Biofuels 2009, 11, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koutsandreas, T.; Ladoukakis, E.; Pilalis, E.; Zarafeta, D.; Kolisis, F.N.; Skretas, G.; Chatziioannou, A.A. ANASTASIA: An Automated Metagenomic Analysis Pipeline for Novel Enzyme Discovery Exploiting next Generation Sequencing Data. Front. Genet. 2019, 10, 469. [Google Scholar] [CrossRef] [PubMed]

| Enzyme | Source | Special Feature/Screening Method | Application | References |
|---|---|---|---|---|
| Cellulase | Buffalo rumen | Thermotolerant | Bioethanol, biopolishing of textile fibers | [17] |
| Cellulase | Anaerobic beer | Halotolerant | Bioethanol, biodiesel, extraction of flavoring materials and essential oils | [18] |
| Hemicellulase | Degrading wheat straw | Thermotolerant, halotolerant | Bioethanol, biodiesel, bioplastic from wood biomass | [19] |
| Xylanase | Sea-bottom | Sequence based | Bioethanol | [20] |
| Xyloglucanase | Bovine rumen | Activity based | Bioethanol | [21] |
| Acidobacterial-laccase like copper oxidase | Marsh bog soil | Thermostable, halotolerant, stable in the presence of organic solvents | Polyphenol removal from wine, bioethanol production | [22] |
| Esterase | Antarctic soil | Function based (fosmid vector) | Bioethanol production | [23] |
| Glycoside hydrolases | Anaerobic digestion sludge | Sequence based | Bioethanol production | [24] |
| Cyclodextrinase | Bovine rumen | Function based | Biodiesel production | [25] |
| Enzymes involved in bioremediation | ||||
| Naphthalene dioxygenase | Oil-contaminated soil | Function based | Bioremediation of oil-contaminated soil/water | [26] |
| Oxygenases | Artificially polluted soil | Function based | Remediation of oil-contaminated soil/water | [27] |
| Laccase | Water from South China Sea | Sequencing based | Dye degradation | [28] |
| Phenol hydroxylases and catechol 2,3-dioxygenases | Wastewater treatment plant | Function based | Potential use in aromatic compound degradation | [29] |
| Enzymes used in other applications | ||||
| Lipase/protease/hemolysins/biosurfactants | Slaughterhouse drain | Function based | Applied as antimicrobial agent | [30] |
| β-Glucosidase | Hydrothermal spring water | Function based | Thermotolerant and heath-active enzyme | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sartaj, K.; Patel, A.; Matsakas, L.; Prasad, R. Unravelling Metagenomics Approach for Microbial Biofuel Production. Genes 2022, 13, 1942. https://doi.org/10.3390/genes13111942
Sartaj K, Patel A, Matsakas L, Prasad R. Unravelling Metagenomics Approach for Microbial Biofuel Production. Genes. 2022; 13(11):1942. https://doi.org/10.3390/genes13111942
Chicago/Turabian StyleSartaj, Km, Alok Patel, Leonidas Matsakas, and Ramasare Prasad. 2022. "Unravelling Metagenomics Approach for Microbial Biofuel Production" Genes 13, no. 11: 1942. https://doi.org/10.3390/genes13111942