Potassium Transporter LrKUP8 Is Essential for K+ Preservation in Lycium ruthenicum, A Salt-Resistant Desert Shrub
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material for Ion Content Analysis
2.2. Determination of Na+ and K+ Contents
2.3. Callus Induction, Selection, and Maintenance
2.4. Isolation of Putative Key Genes Responsible for K+ /Na+ Homeostasis
2.5. Subcellular Localization of LrKUP8
2.6. Gene Expression Analysis by Quantitative Real-Time PCR
2.7. Agrobacterium-Mediated Transformation of L. ruthenicum Calli
2.8. Observation of GFP Fluorescence
2.9. Determination of Callus Growth under Salt Stress
2.10. Ion Contents
2.11. Ion Flux
2.12. Statistical Analysis
3. Results
3.1. K+ Distribution under Normal and Salinized Conditions
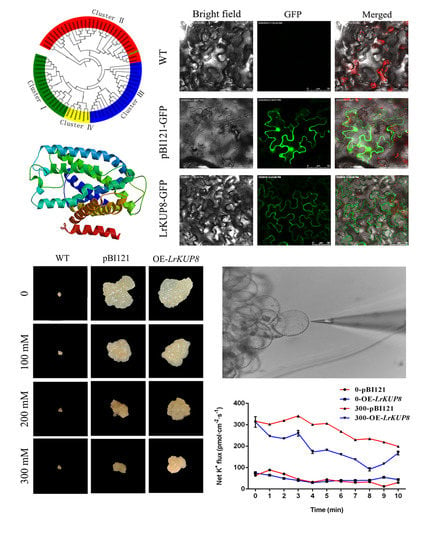
3.2. Cloning, Protein Sequence Alignment, and Phylogenetic Analysis of LrKUP8
3.3. The Subcellular Localization and Expression Pattern of LrKUP8
3.4. Overexpression of LrKUP8 Improved Callus Salt Tolerance
3.5. LrKUP8 Functions in K+/Na+ Homeostasis through K Accumulation under Salt Stress Condition
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DW | dry weight |
| FW | fresh weight |
| MDA | malondialdehyde |
| MS | Murashige and Skoog |
| NMT | non-invasive micro-test technique |
| PM | plasma membrane |
| qPCR | quantitative real-time PCR |
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Glenn, E.P.; Volkov, V. Could vesicular transport of Na+ and Cl− be a feature of salt tolerance in halophytes? Ann. Bot. 2019, 123, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Ansari, R.; Ali, H.; Gul, B.; Nielsen, B.L. Panicum turgidum, a potentially sustainable cattle feed alternative to maize for saline areas. Agric. Ecosyst. Environ. 2009, 129, 542–546. [Google Scholar] [CrossRef]
- Koyro, H.W.; Zörb, C.; Debez, A.; Huchzermeyer, B. The effect of hyperosmotic salinity on protein pattern and enzyme activities of halophytes. Funct. Plant Biol. 2013, 40, 787–804. [Google Scholar] [CrossRef]
- Jin, H.; Liu, Y.; Yang, F.; Wang, J.; Fu, D.; Zhang, X. Characterization of anthocyanins in wild Lycium ruthenicum Murray by HPLC-DAD/QTOF-MS/MS. Anal. Methods 2015, 7, 4947–4956. [Google Scholar] [CrossRef]
- Ali, A.; Khan, I.U.; Jan, M.; Khan, H.A.; Hussain, S.; Nisar, M.; Chung, W.S.; Yun, D.-J. The high-affinity Potassium transporter EpHKT1;2 from the extremophile Eutrema parvula mediates salt tolerance. Front. Plant Sci. 2018, 9, 1108. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Yun, D.-J. Differential selection of sodium and potassium ions by TsHKT1;2. Plant Signal. Behav. 2016, 11, e1206169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, S.; Ma, X.; Zhu, H.; Sa, G.; Sun, J.; Li, N.; Zhao, C.; Zhao, R.; Chen, S. Extracellular ATP mediates cellular K+/Na+ homeostasis in two contrasting poplar species under NaCl stress. Trees 2016, 30, 825–837. [Google Scholar] [CrossRef]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. Hkt transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 2015, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Z.; Zhou, Y.; Han, J.; Shi, D. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.H. Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Véry, A.A.; Sentenac, H. Molecular Mechanisms and Regulation of K+ Transport in Higher Plants. Annu. Rev. Plant Biol. 2003, 54, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Grabov, A. Plant KT/KUP/HAK potassium transporters: Single family-multiple functions. Ann. Bot. 2007, 99, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Qiu, X.H.; Wang, L.; Xie, W.B.; Zhang, C.J.; Xiong, L.Z.; Lian, X.M.; Zhang, Q.F. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genom. 2008, 280, 437–452. [Google Scholar] [CrossRef]
- Qi, Z.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol. Plant 2010, 3, 326–333. [Google Scholar] [CrossRef]
- Horie, T.; Sugawara, M.; Okada, T.; Taira, K.; Kaothien-Nakayama, P.; Katsuhara, M.; Nakayama, H. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 2011, 111, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Xu, G. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gao, Q.; Sun, C.; Li, W.; Gu, S.; Xu, C. Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). J. Genet. Genom. 2009, 36, 161–172. [Google Scholar] [CrossRef]
- Fu, H.H.; Luan, S. AtKUP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 1998, 10, 63–73. [Google Scholar] [PubMed]
- Elumalai, R.P.; Nagpal, P.; Reed, J.W. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 2002, 14, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Agullo, F.; Rigas, S.; Desbrosses, G.; Dolan, L.; Hatzopoulos, P.; Grabov, A. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 2004, 40, 523–535. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, L.; Li, Q.; Jiang, Z.; Liu, T.; Zhu, Y. The effects of exogenous antioxidant germanium (Ge) on seed germination and growth of Lycium ruthenicum Murr subjected to NaCl stress. Environ. Technol. 2016, 37, 909–919. [Google Scholar] [CrossRef]
- Wang, C.M.; Xia, C.M.; Wu, G.Q.; Yuan, H.J.; Wang, X.R.; Li, J.H.; Tian, F.P.; Zhang, Q.; Wang, H.; Li, J.; et al. Lycium ruthenicum studies: Molecular biology, phytochemistry and pharmacology. Food Chem. 2018, 240, 759–766. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Zhang, W. Study on hypoglycemic function of polysaccharides from Lycium ruthenicum Murr. fruit and its mechanism. Food Sci. 2009, 30, 244–248. [Google Scholar]
- Peng, Q.; Lv, X.; Xu, Q.; Li, Y.; Huang, L.; Du, Y. Isolation and structural characterization of the polysaccharide LRGP1 from Lycium ruthenicum. Carbohyd. Polym. 2012, 90, 95–101. [Google Scholar] [CrossRef]
- Peng, Q.; Liu, H.; Shi, S.; Li, M. Lycium ruthenicum polysaccharide attenuates inflammation through inhibiting TLR4/NF-κ B signaling pathway. Int. J. Biol. Macromol. 2014, 67, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dang, J.; Wang, Q.; Yu, M.; Jiang, L.; Mei, L.; Shao, Y.; Tao, Y. Optimization of polysaccharides from Lycium ruthenicum fruit using RSM and its anti-oxidant activity. Int. J. Biol. Macromol. 2013, 61, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, Y.; Zeng, S.; Patra, B.; Yuan, L.; Wang, Y. Isolation and characterization of a salt stress-responsive betaine aldehyde dehydrogenase in Lycium ruthenicum Murr. Physiol. Plant 2018, 163, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ding, C.; Wang, L.; Li, G.; Shi, J.; Li, H.; Wang, H.; Suo, Y. Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Food Chem. 2011, 126, 859–865. [Google Scholar] [CrossRef]
- Walsh, A. The application of atomic absorption spectra to chemical analysis. Spectrochim. Acta 1955, 7, 108–117. [Google Scholar] [CrossRef]
- Chen, J.H.; Zhang, D.Z.; Zhang, C.; Xu, M.L.; Yin, W.L. Physiological characterization, transcriptomic profiling, and microsatellite marker mining of Lycium ruthenicum. J. Zhejiang Univ. Sci B 2017, 18, 1002–1021. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, Y.; Wu, M.; Liu, X.; Shen, X.; Liu, C.; Wang, Y. Identification and validation of reference genes for quantitative real-Time PCR normalization and its applications in Lycium. PLoS ONE 2014, 9, e97039. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Schm method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 2009, 149, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.S.; Gao, H.; Sun, S. The salt resistance of wild soybean (Glycine soja Sieb. et Zucc. ZYD 03262) under NaCl stress is mainly determined by Na+ distribution in the plant. Acta. Physiol. Plant 2014, 36, 61–70. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, X.M.; Fan, C.M.; Chen, F.L.; Zhu, J.L.; Zhang, S.L.; Chen, Q.S.; Fu, Y.F. A callus transformation system for gene functional studies in soybean. J. Integr. Agric. 2017, 16, 1913–1922. [Google Scholar] [CrossRef] [Green Version]
- Chahel, A.A.; Zeng, S.; Yousaf, Z.; Liao, Y.; Yang, Z.; Wei, X.; Ying, W. Plant-specific transcription factor LrTCP4 enhances secondary metabolite biosynthesis in Lycium ruthenicum hairy roots. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 136, 323–337. [Google Scholar] [CrossRef]
- Ismail, H.; Maksimović, J.D.; Maksimović, V.; Shabala, L.; Živanović, B.D.; Tian, Y.; Jacobsen, S.E.; Shabala, S. Rutin, a flavonoid with antioxidant activity, improves plant salinity tolerance by regulating K+ retention and Na+ exclusion from leaf mesophyll in quinoa and broad beans. Funct. Plant Biol. 2016, 43, 75–86. [Google Scholar] [CrossRef]
- Wang, N.; Qi, H.K.; Qiao, W.Q.; Shi, J.B.; Xu, Q.H.; Zhou, H.; Yan, G.T.; Huang, Q. Cotton (Gossypium hirsutum L.) genotypes with contrasting K+/Na+ ion homeostasis: Implications for salinity tolerance. Acta Physiol. Plant 2017, 39, 77. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Wakeel, A.; Faroop, M.; Qadir, M.; Schubert, S. Potassium substitution by sodium in plants. Crit. Rev. Plant Sci. 2011, 30, 401–413. [Google Scholar] [CrossRef]
- Zhu, X.Q.; He, J.J.; Kumar, T.; Wang, X.L.; Zhang, J.L. The coordinated regulation of Na+ and K+ in Hordeum brevisubulatum responding to time of salt stress. Plant Sci. 2016, 252, 358–366. [Google Scholar]
- Wu, G.Q.; Wang, J.L.; Li, S.J. Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar Beet (β vulgaris L.) and their regulated expression under salt stress. Genes 2019, 10, 401. [Google Scholar] [CrossRef]
- Colmer, T.D.; Flowers, T.J.; Munns, R. Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 2006, 57, 1059–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuin, T.A.; Miller, A.J.; Laurie, S.A.; Leigh, R.A. Potassium activities in cell compartments of salt-grown barley leaves. J. Exp. Bot. 2003, 54, 657–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gierth, M.; Maser, P. Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007, 58, 2348–2356. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.L.; Rogers, M.E. Arguments for the use of physiological criteria for improving the salt tolerance in crops. Plant Soil 1992, 146, 99–107. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Klei, K.; Fokkens, L.; Haring, M.A.; Schranz, M.E.; Testerink, C. Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exp. Bot. 2016, 67, 2127–2138. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Nishio, T.; Ichizen, N.; Takano, T. High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Rep. 2007, 26, 1673–1679. [Google Scholar] [CrossRef]
- Shabala, L.; Cuin, T.A.; Newman, I.A.; Shabala, S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 2005, 222, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.S. Non-invasive ion probes-tools for measuring transmembrane ion flux. Nature 1995, 378, 645–646. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, F.; Li, A.; Rao, S.; Chen, J. Potassium Transporter LrKUP8 Is Essential for K+ Preservation in Lycium ruthenicum, A Salt-Resistant Desert Shrub. Genes 2019, 10, 600. https://doi.org/10.3390/genes10080600
Dai F, Li A, Rao S, Chen J. Potassium Transporter LrKUP8 Is Essential for K+ Preservation in Lycium ruthenicum, A Salt-Resistant Desert Shrub. Genes. 2019; 10(8):600. https://doi.org/10.3390/genes10080600
Chicago/Turabian StyleDai, Fengbin, Aijia Li, Shupei Rao, and Jinhuan Chen. 2019. "Potassium Transporter LrKUP8 Is Essential for K+ Preservation in Lycium ruthenicum, A Salt-Resistant Desert Shrub" Genes 10, no. 8: 600. https://doi.org/10.3390/genes10080600