Rho GTPases in the Amygdala—A Switch for Fears?
Abstract
:1. Introduction
2. The Amygdala Is the Fear Center in the Brain
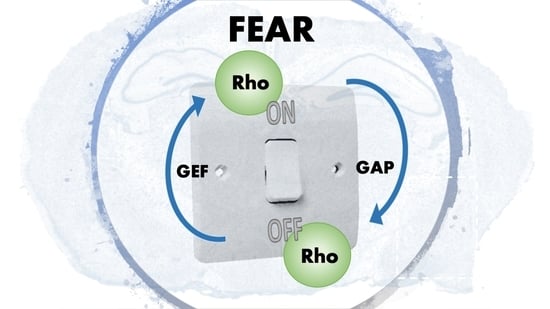
3. Rho GTPases and Their Regulation
4. Rho GTPases in Synaptic Spine Plasticity
5. Rho GTPases and Fear Conditioning in the Amygdala
6. The Critical Role of Rich2 in Fear Modulation
6.1. Rich2 Deletion Induces Specific Behavioral Changes in Mice
6.2. Rich2 Deactivates Different Rho GTPases in Different Brain Regions
6.3. Spine Morphology in Rich2 Knock-Out Animals
6.4. Deletion of Rich2 Affects the Immediate-Early Gene Expression in the Amygdala
6.5. Shank3-Rich2 Interaction in the Context of Fear Learning
7. Rho GTPases and Fear Modulation in Neurological/Neuropsychiatric Disorders with Anxiety and Phobia as Comorbidities
7.1. Rho GTPases in Mood Disorders
7.2. Rho GTPases in Autism Spectrum Disorders
7.3. Rho GTPases in Schizophrenia (SCZ)
7.4. Rho GTPases in Intellectual Disability (ID)
8. Rho GTPases as a Potential Drug Target
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aggleton, J.P. The Amygdala: A Functional Analysis; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Calhoon, G.G.; Tye, K.M. Resolving the neural circuits of anxiety. Nat. Neurosci. 2015, 18, 1394–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, L.M.; Rauch, S.L.; Pitman, R.K. Amygdala, Medial Prefrontal Cortex, and Hippocampal Function in PTSD. Ann. N. Y. Acad. Sci. 2006, 1071, 67–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, B.M.; Milad, M.R. The Study of Fear Extinction: Implications for Anxiety Disorders. Am. J. Psychiatry 2011, 168, 1255–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilbronner, S.R.; Rodríguez-Romaguera, J.; Quirk, G.J.; Groenewegen, H.J.; Haber, J.E. Circuit-Based Corticostriatal Homologies Between Rat and Primate. Biol. Psychiatry 2016, 80, 509–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cryan, J.F.; Sweeney, F.F. The age of anxiety: Role of animal models of anxiolytic action in drug discovery. Br. J. Pharmacol. 2011, 164, 1129–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gafford, G.M.; Ressler, K.J. Mouse models of fear-related disorders: Cell-type-specific manipulations in amygdala. Neuroscience 2015, 321, 108–120. [Google Scholar] [CrossRef] [Green Version]
- File, S.; Gonzalez, L.; Gallant, R. Role of the Basolateral Nucleus of the Amygdala in the Formation of a Phobia. Neuropsychopharmacology 1998, 19, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Mahan, A.L.; Ressler, K.J. Fear conditioning, synaptic plasticity and the amygdala: Implications for posttraumatic stress disorder. Trends Neurosci. 2012, 35, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Markram, K.; Rinaldi, T.; La Mendola, D.; Sandi, C.; Markram, H. Abnormal Fear Conditioning and Amygdala Processing in an Animal Model of Autism. Neuropsychopharmacology 2007, 33, 901–912. [Google Scholar] [CrossRef]
- Philippe, A.; Boddaert, N.; Vaivre-Douret, L.; Robel, L.; Danon-Boileau, L.; Malan, V.; De Blois, M.-C.; Heron, D.; Colleaux, L.; Golse, B.; et al. Neurobehavioral Profile and Brain Imaging Study of the 22q13.3 Deletion Syndrome in Childhood. Pediatrics 2008, 122, e376–e382. [Google Scholar] [CrossRef]
- Gray, J.L.; Von Delft, F.; Brennan, P. Targeting the Small GTPase Superfamily through Their Regulatory Proteins. Angew. Chem. Int. Ed. 2020, 59, 6342–6366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, D.R.; Rossman, K.L.; Der, C.J. Rho guanine nucleotide exchange factors: Regulators of Rho GTPase activity in development and disease. Oncogene 2013, 33, 4021–4035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, D.; Yang, X.; Shi, L. Rho GTPase Regulators and Effectors in Autism Spectrum Disorders: Animal Models and Insights for Therapeutics. Cells 2020, 9, 835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niftullayev, S.; Lamarche-Vane, N. Regulators of Rho GTPases in the Nervous System: Molecular Implication in Axon Guidance and Neurological Disorders. Int. J. Mol. Sci. 2019, 20, 1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamboni, V.; Jones, R.; Umbach, A.; Ammoni, A.; Passafaro, M.; Hirsch, E.; Merlo, G.R. Rho GTPases in Intellectual Disability: From Genetics to Therapeutic Opportunities. Int. J. Mol. Sci. 2018, 19, 1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsini, C.A.; Maren, S. Neural and cellular mechanisms of fear and extinction memory formation. Neurosci. Biobehav. Rev. 2012, 36, 1773–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fanselow, M.S.; Poulos, A.M. The Neuroscience of Mammalian Associative Learning. Annu. Rev. Psychol. 2005, 56, 207–234. [Google Scholar] [CrossRef] [Green Version]
- Krettek, J.E.; Price, J.L. A description of the amygdaloid complex in the rat and cat with observations on intra-amygdaloid axonal connections. J. Comp. Neurol. 1978, 178, 255–279. [Google Scholar] [CrossRef]
- Spampanato, J.; Polepalli, J.S.; Sah, P. Interneurons in the basolateral amygdala. Neuropharmacology 2011, 60, 765–773. [Google Scholar] [CrossRef]
- Poulin, J.-F.; Laforest, S.; Drolet, G.; Castonguay-Lebel, Z. Enkephalin co-expression with classic neurotransmitters in the amygdaloid complex of the rat. J. Comp. Neurol. 2008, 506, 943–959. [Google Scholar] [CrossRef]
- Benham, R.S.; Engin, E.; Rudolph, U. Diversity of Neuronal Inhibition. JAMA Psychiatry 2014, 71, 91–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvarci, S.; Paré, D. Amygdala Microcircuits Controlling Learned Fear. Neuron 2014, 82, 966–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ressler, R.L.; Maren, S. Synaptic encoding of fear memories in the amygdala. Curr. Opin. Neurobiol. 2019, 54, 54–59. [Google Scholar] [CrossRef]
- Pape, H.-C.; Pare, D. Plastic synaptic networks of the amygdala for the acquisition, expression, and extinction of conditioned fear. Physiol. Rev. 2010, 90, 419–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, J.P.; Hamanaka, H.; Monfils, M.-H.; Behnia, R.; Deisseroth, K.; Blair, H.T.; LeDoux, J.E. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc. Natl. Acad. Sci. USA 2010, 107, 12692–12697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, J.-T.; Nakajima, R.; Kim, H.-S.; Jeong, Y.; Augustine, G.J.; Han, J.-H. Optogenetic activation of presynaptic inputs in lateral amygdala forms associative fear memory. Learn. Mem. 2014, 21, 627–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabavi, S.; Fox, R.; Proulx, C.D.; Lin, J.Y.; Tsien, R.Y.; Malinow, R. Engineering a memory with LTD and LTP. Nature 2014, 511, 348–352. [Google Scholar] [CrossRef]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Bishop, A.L.; Hall, A. Rho GTPases and their effector proteins. Biochem. J. 2000, 348, 241–255. [Google Scholar] [CrossRef]
- Vetter, I.R.; Wittinghofer, A. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef] [Green Version]
- Cherfils, J.; Zeghouf, M. Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, E. Deciphering the molecular and functional Basis of RHOGAP family proteins A systematic approach toward selective inactivation of rho family proteINS. J. Biol. Chem. 2016, 291, 20353–20371. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DerMardirossian, C.; Bokoch, G.M. GDIs: Central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005, 15, 356–363. [Google Scholar] [CrossRef]
- Wherlock, M.; Mellor, H. The Rho GTPase family: A Racs to Wrchs story. J. Cell Sci. 2002, 115, 239–240. [Google Scholar]
- Luo, L. RHO GTPASES in neuronal morphogenesis. Nat. Rev. Neurosci. 2000, 1, 173–180. [Google Scholar] [CrossRef]
- Zhang, H. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 2005, 25, 3379–3388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maekawa, M. Signaling from Rho to the Actin Cytoskeleton Through Protein Kinases ROCK and LIM-kinase. Science 1999, 285, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Engert, F.; Bonhoeffer, T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 1999, 399, 66–70. [Google Scholar] [CrossRef]
- Nimchinsky, E.A.; Sabatini, B.L.; Svoboda, K. Structure and Function of Dendritic Spines. Annu. Rev. Physiol. 2002, 64, 313–353. [Google Scholar] [CrossRef] [Green Version]
- Harris, K.M. Calcium from internal stores modifies dendritic spine shape. Proc. Natl. Acad. Sci. USA 1999, 96, 12213–12215. [Google Scholar] [CrossRef] [Green Version]
- Penzes, P.; Cahill, M.E.; Jones, K.; VanLeeuwen, J.-E.; Woolfrey, K.M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011, 14, 285–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourne, J.; Harris, K.M. Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 2008, 31, 47–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, M.L.; Armstrong, N. Structure and Function of Glutamate Receptor Ion Channels. Annu. Rev. Physiol. 2004, 66, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Lerma, J. Roles and rules of kainate receptors in synaptic transmission. Nat. Rev. Neurosci. 2003, 4, 481–495. [Google Scholar] [CrossRef]
- Sarowar, T.; Grabrucker, A.M. Actin-Dependent Alterations of Dendritic Spine Morphology in Shankopathies. Neural Plast. 2016, 2016, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, A.; Minden, A.; Yuste, R. Regulation of dendritic spine morphology by the rho family of small GTPases: Antagonistic roles of Rac and Rho. Cereb. Cortex 2000, 10, 927–938. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, A.Y.; Harms, M.B.; Luo, L. Small GTPases Rac and Rho in the Maintenance of Dendritic Spines and Branches in Hippocampal Pyramidal Neurons. J. Neurosci. 2000, 20, 5329–5338. [Google Scholar] [CrossRef]
- Bongmba, O. The Role of Rac1 in Synaptic Plasticity, Learning and Memory; University of Houston: Houston, TX, USA, 2013. [Google Scholar]
- Martinez, L.A.; Tejada-Simon, M.V. Pharmacological inactivation of the small GTPase Rac1 impairs long-term plasticity in the mouse hippocampus. Neuropharmacology 2011, 61, 305–312. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.H.; Wang, H.; Soderling, S.H.; Yasuda, R. Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. eLife 2014, 3, e02839. [Google Scholar] [CrossRef]
- Firat-Karalar, E.N.; Welch, M.D. New mechanisms and functions of actin nucleation. Curr. Opin. Cell Biol. 2011, 23, 4–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shekhar, S.; Pernier, J.; Carlier, M.-F. Regulators of actin filament barbed ends at a glance. J. Cell Sci. 2016, 129, 1085–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorra, K.E.; Harris, K.M. Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus 2000, 10, 501–511. [Google Scholar] [CrossRef]
- Okamoto, K.-I.; Nagai, T.; Miyawaki, A.; Hayashi, Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 2004, 7, 1104–1112. [Google Scholar] [CrossRef]
- Huganir, R.L.; Nicoll, R.A. AMPARs and synaptic plasticity: The last 25 years. Neuron 2013, 80, 704–717. [Google Scholar] [CrossRef] [Green Version]
- Casey, P.J.; Seabra, M.C. Protein Prenyltransferases. J. Biol. Chem. 1996, 271, 5289–5292. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.-P.; Wu, K.-Y.; Liang, B.; Fu, X.-Q.; Luo, Z.-G. TrkB-mediated activation of geranylgeranyltransferase I promotes dendritic morphogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 17181–17186. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Gao, S.; Sun, C.; Chen, L.; Shi, Q.; Hu, J.; Yu, R.; Zhou, X. Inhibiting geranylgeranyltransferase I activity decreases spine density in central nervous system. Hippocampus 2014, 25, 373–384. [Google Scholar] [CrossRef]
- Fiala, J.C.; Spacek, J.; Harris, K.M. Dendritic Spine Pathology: Cause or Consequence of Neurological Disorders? Brain Res. Rev. 2002, 39, 29–54. [Google Scholar] [CrossRef]
- Nestler, E.J. Molecular basis of long-term plasticity underlying addiction. Nat. Rev. Neurosci. 2001, 2, 119–128. [Google Scholar] [CrossRef]
- Hutsler, J.J.; Zhang, H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010, 1309, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.E.; Hennessey, T.M.; Gordon, K.M.; Ryan, S.J.; McNair, M.L.; Ressler, K.J.; Rainnie, D.G. Developmental disruption of amygdala transcriptome and socioemotional behavior in rats exposed to valproic acid prenatally. Mol. Autism 2017, 8, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kõks, S.; Luuk, H.; Nelovkov, A.; Areda, T.; Vasar, E. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav. 2004, 3, 80–89. [Google Scholar] [CrossRef]
- Hou, Y.-Y.; Lu, B.; Li, M.; Liu, Y.; Chen, J.; Chi, Z.-Q.; Liu, J.-G. Involvement of Actin Rearrangements within the Amygdala and the Dorsal Hippocampus in Aversive Memories of Drug Withdrawal in Acute Morphine-Dependent Rats. J. Neurosci. 2009, 29, 12244–12254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhou, Q.-X.; Hou, Y.-Y.; Lu, B.; Yu, C.; Chen, J.; Ling, Q.-L.; Cao, J.; Chi, Z.-Q.; Xu, L.; et al. Actin Polymerization-Dependent Increase in Synaptic Arc/Arg3.1 Expression in the Amygdala Is Crucial for the Expression of Aversive Memory Associated with Drug Withdrawal. J. Neurosci. 2012, 32, 12005–12017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, K.T.; Monsey, M.S.; Wu, M.S.; Schafe, G.E. Synaptic Plasticity and NO-cGMP-PKG Signaling Regulate Pre- and Postsynaptic Alterations at Rat Lateral Amygdala Synapses Following Fear Conditioning. PLoS ONE 2010, 5, e11236. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, C.; Wang, R.; Govindarajan, S.; Barish, P.; Vernon, M.; Fu, C.; Acharya, A.; Chen, L.; Boykin, E.; et al. Corticosterone induced morphological changes of hippocampal and amygdaloid cell lines are dependent on 5-HT7 receptor related signal pathway. Neuroscience 2011, 182, 71–81. [Google Scholar] [CrossRef]
- Martinez, L.A.; Klann, E.; Tejada-Simon, M.V. Translocation and activation of Rac in the hippocampus during associative contextual fear learning. Neurobiol. Learn. Mem. 2007, 88, 104–113. [Google Scholar] [CrossRef]
- Sananbenesi, F.; Fischer, A.; Wang, X.; Schrick, C.; Neve, R.; Radulovic, J.; Tsai, L.-H. A hippocampal Cdk5 pathway regulates extinction of contextual fear. Nat. Neurosci. 2007, 10, 1012–1019. [Google Scholar] [CrossRef] [Green Version]
- Diana, G.; Valentini, G.; Travaglione, S.; Falzano, L.; Pieri, M.; Zona, C.; Meschini, S.; Fabbri, A.; Fiorentini, C. Enhancement of learning and memory after activation of cerebral Rho GTPases. Proc. Natl. Acad. Sci. USA 2007, 104, 636–641. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Ding, Z.-B.; Meng, S.-Q.; Shen, H.-W.; Sun, S.-C.; Luo, Y.-X.; Liu, J.-F.; Lu, L.; Zhu, W.-L.; Shi, J. Differential role of Rac in the basolateral amygdala and cornu ammonis 1 in the reconsolidation of auditory and contextual Pavlovian fear memory in rats. Psychopharmacology 2014, 231, 2909–2919. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yao, W.; Wang, J.; Yang, T.; Liu, C.; Tao, Y.; Chen, Y.; Liu, X.; Ma, L. Post-training activation of Rac1 in the basolateral amygdala is required for the formation of both short-term and long-term auditory fear memory. Front. Mol. Neurosci. 2015, 8, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Dines, M.; Alapin, J.M.; Lamprecht, R. Affecting long-term fear memory formation through optical control of Rac1 GTPase and PAK activity in lateral amygdala. Sci. Rep. 2017, 7, 13930. [Google Scholar] [CrossRef]
- Wang, W.-S.; Ju, Y.-Y.; Zhou, Q.-X.; Tang, J.-X.; Li, M.; Zhang, L.; Kang, S.; Chen, Z.-G.; Wang, Y.-J.; Ji, H.; et al. The Small GTPase Rac1 Contributes to Extinction of Aversive Memories of Drug Withdrawal by Facilitating GABAA Receptor Endocytosis in the vmPFC. J. Neurosci. 2017, 37, 7096–7110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Z.; Tao, Y.; Guo, X.; Cheng, D.; Wang, F.; Liu, X.; Ma, L. Fear Conditioning Downregulates Rac1 Activity in the Basolateral Amygdala Astrocytes to Facilitate the Formation of Fear Memory. Front. Mol. Neurosci. 2017, 10, 396. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.M.; Johnson, R.C.; Mains, R.; Eipper, B. Expression of kalirin, a neuronal GDP/GTP exchange factor of the trio family, in the central nervous system of the adult rat. J. Comp. Neurol. 2001, 429, 388–402. [Google Scholar] [CrossRef]
- Sarowar, T.; Grabrucker, S.; Boeckers, T.M.; Grabrucker, A.M. Object Phobia and Altered RhoA Signaling in Amygdala of Mice Lacking Rich2. Front. Mol. Neurosci. 2017, 10, 180. [Google Scholar] [CrossRef] [Green Version]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 22 August 2020).
- Kato, T.; Ishiwata, M.; Yamada, K.; Kasahara, T.; Kakiuchi, C.; Iwamoto, K.; Kawamura, K.; Ishihara, H.; Oka, Y. Behavioral and gene expression analyses of Wfs1 knockout mice as a possible animal model of mood disorder. Neurosci. Res. 2008, 61, 143–158. [Google Scholar] [CrossRef]
- Nasu-Nishimura, Y.; Hayashi, T.; Ohishi, T.; Okabe, T.; Ohwada, S.; Hasegawa, Y.; Senda, T.; Toyoshima, C.; Nakamura, T.; Akiyama, T. Role of the Rho GTPase-activating protein RICS in neurite outgrowth. Genes Cells 2006, 11, 607–614. [Google Scholar] [CrossRef]
- Levy, R.J.; Kvajo, M.; Li, Y.; Tsvetkov, E.; Dong, W.; Yoshikawa, Y.; Kataoka, T.; Bolshakov, V.Y.; Karayiorgou, M.; Gogos, J.A. Deletion of Rapgef6, a candidate schizophrenia susceptibility gene, disrupts amygdala function in mice. Transl. Psychiatry 2015, 5, e577. [Google Scholar] [CrossRef]
- Govek, E.-E.; Newey, S.E.; Akerman, C.J.; Cross, J.R.; Van Der Veken, L.; Van Aelst, L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 2004, 7, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Endris, V.; Wogatzky, B.; Leimer, U.; Bartsch, D.; Zatyka, M.; Latif, F.; Maher, E.; Tariverdian, G.; Kirsch, S.; Karch, D.; et al. The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc. Natl. Acad. Sci. USA 2002, 99, 11754–11759. [Google Scholar] [CrossRef] [Green Version]
- Sarowar, T.; Grabrucker, S.; Föhr, K.; Mangus, K.; Eckert, M.; Bockmann, J.; Boeckers, T.M.; Grabrucker, A.M. Enlarged dendritic spines and pronounced neophobia in mice lacking the PSD protein RICH2. Mol. Brain 2016, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarowar, T. Characterization of RICH2 Knock-Out Mouse Model; Universität Ulm: Ulm, Germany, 2017. [Google Scholar]
- Dahl, J. Biochemical and Functional Characterization of RhoSAP: A RhoGAP of the Postsynaptic Density; Universität Ulm: Ulm, Germany, 2009. [Google Scholar]
- Madani, R.; Kozlov, S.; Akhmedov, A.; Cinelli, P.; Kinter, J.; Lipp, H.-P.; Sonderegger, P.; Wolfer, D.P. Impaired explorative behavior and neophobia in genetically modified mice lacking or overexpressing the extracellular serine protease inhibitor neuroserpin. Mol. Cell. Neurosci. 2003, 23, 473–494. [Google Scholar] [CrossRef]
- Abe, K.; Rossman, K.L.; Liu, B.; Ritola, K.D.; Chiang, D.; Campbell, S.L.; Burridge, K.; Der, C.J. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 2000, 275, 10141–10149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richnau, N.; Aspenström, P. RICH, a Rho GTPase-activating Protein Domain-containing Protein Involved in Signaling by Cdc42 and Rac1. J. Biol. Chem. 2001, 276, 35060–35070. [Google Scholar] [CrossRef] [Green Version]
- Ridgway, L.D.; Wetzel, M.D.; Marchetti, D. Modulation of GEF-H1 Induced Signaling by Heparanase in Brain Metastatic Melanoma Cells. J. Cell. Biochem. 2010, 111, 1299–1309. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.P.; Burridge, K. Vav2 Activates Rac1, Cdc42, and RhoA Downstream from Growth Factor Receptors but Not β1 Integrins. Mol. Cell. Biol. 2000, 20, 7160–7169. [Google Scholar] [CrossRef] [Green Version]
- Raynaud, F. Rho-GTPase-activating Protein Interacting with Cdc-42-interacting Protein 4 Homolog 2 (Rich2) A NEW Ras-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 (Rac1) GTPase-ACTIVATING PROTEIN THAT CONTROLS DENDRITIC SPINE MORPHOGENESIS. J. Biol Chem. 2014, 289, 2600–2609. [Google Scholar] [CrossRef] [Green Version]
- Guilluy, C.; Garcia-Mata, R.; Burridge, K. Rho protein crosstalk: Another social network? Trends Cell Biol. 2011, 21, 718–726. [Google Scholar] [CrossRef] [Green Version]
- Newey, S.E.; Velamoor, V.; Govek, E.-E.; Van Aelst, L. Rho GTPases, dendritic structure, and mental retardation. J. Neurobiol. 2005, 64, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P. BAR domain proteins regulate Rho GTPase signaling. Small GTPases 2014, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Disanza, A.; Carlier, M.-F.; Stradal, T.E.B.; Didry, M.; Frittoli, E.; Confalonieri, S.; Croce, A.; Wehland, J.; Di Fiore, P.P.; Scita, G. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat. Cell Biol. 2004, 6, 1180–1188. [Google Scholar] [CrossRef]
- Ueyama, T.; Sakoda, T.; Kawashima, S.; Hiraoka, E.; Hirata, K.-I.; Akita, H.; Yokoyama, M. Activated RhoA stimulates c-fos gene expression in myocardial cells. Circ. Res. 1997, 81, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Bozdagi-Gunal, O.; Sakurai, T.; Papapetrou, D.; Wang, X.; Dickstein, D.L.; Takahashi, N.; Kajiwara, Y.; Yang, M.; Katz, A.M.; Scattoni, M.L.; et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol. Autism 2010, 1, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, Y.-E.; Yoo, T.; Lee, S.; Lee, J.; Kim, D.; Han, H.-M.; Bae, Y.-C.; Kim, E. Shank3 Mice Carrying the Human Q321R Mutation Display Enhanced Self-Grooming, Abnormal Electroencephalogram Patterns, and Suppressed Neuronal Excitability and Seizure Susceptibility. Front. Mol. Neurosci. 2019, 12, 155. [Google Scholar] [CrossRef]
- Yang, M.; Bozdagi, O.; Scattoni, M.L.; Wöhr, M.; Roullet, F.I.; Katz, A.M.; Abrams, D.N.; Kalikhman, D.; Simon, H.; Woldeyohannes, L.; et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J. Neurosci. 2012, 32, 6525–6541. [Google Scholar] [CrossRef]
- Wang, X.; McCoy, P.A.; Rodriguiz, R.M.; Pan, Y.; Je, H.S.; Roberts, A.C.; Kim, C.J.; Berrios, J.; Colvin, J.S.; Bousquet-Moore, D.; et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 2011, 20, 3093–3108. [Google Scholar] [CrossRef] [Green Version]
- Jaramillo, T.C.; Speed, H.E.; Xuan, Z.; Reimers, J.M.; Liu, S.; Powell, C.M. Altered striatal synaptic function and abnormal behaviour in Shank3 exon4-9 deletion mouse model of autism. Autism Res. 2016, 9, 350–375. [Google Scholar] [CrossRef] [Green Version]
- Hulbert, S.W.; Bey, A.L.; Jiang, Y.-H. Environmental enrichment has minimal effects on behavior in the Shank3 complete knockout model of autism spectrum disorder. Brain Behav. 2018, 8, e01107. [Google Scholar] [CrossRef]
- Drapeau, E.; Riad, M.; Kajiwara, Y.; Buxbaum, J.D. Behavioral Phenotyping of an Improved Mouse Model of Phelan-McDermid Syndrome with a Complete Deletion of the Shank3 Gene. Eneuro 2018, 5, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.C.W.; Dodart, J.-C.; Aron, L.; Finley, L.W.; Bronson, R.T.; Haigis, M.C.; Yankner, B.A.; Harper, J.W. Altered social behavior and neuronal development in mice lacking the Uba6-Use1 ubiquitin transfer system. Mol. Cell 2013, 50, 172–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raynaud, F.; Janossy, A.; Dahl, J.; Bertaso, F.; Perroy, J.; Varrault, A.; Vidal, M.; Worley, P.F.; Boeckers, T.M.; Bockaert, J.; et al. Shank3-Rich2 Interaction Regulates AMPA Receptor Recycling and Synaptic Long-Term Potentiation. J. Neurosci. 2013, 33, 9699–9715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarrier, N.; Gregg, L. Suicide risk in civilian PTSD patients. Soc. Psychiatry Psychiatr. Epidemiol. 2004, 39, 655–661. [Google Scholar] [CrossRef]
- Shalev, A.Y.; Rogel-Fuchs, Y.; Pitman, R.K. Conditioned fear and psychological trauma. Biol. Psychiatry 1992, 31, 863–865. [Google Scholar] [CrossRef]
- Deutch, A.; Charney, D.S. A Functional Neuroanatomy of Anxiety and Fear: Implications for the Pathophysiology and Treatment of Anxiety Disorders. Crit. Rev. Neurobiol. 1996, 10, 419–446. [Google Scholar] [CrossRef]
- Hashimoto, R.; Okada, T.; Kato, T.; Kosuga, A.; Tatsumi, M.; Kamijima, K.; Kunugi, H. The Breakpoint Cluster Region Gene on Chromosome 22q11 is Associated with Bipolar Disorder. Biol. Psychiatry 2005, 57, 1097–1102. [Google Scholar] [CrossRef]
- Detera-Wadleigh, S.D.; Liu, C.; Maheshwari, M.; Cardona, I.; Corona, W.; Akula, N.; Steele, C.; Badner, J.A.; Kundu, M.; Kassem, L.; et al. Sequence variation in DOCK9 and heterogeneity in bipolar disorder. Psychiatr. Genet. 2007, 17, 274–286. [Google Scholar] [CrossRef] [Green Version]
- Shi, L. Dock protein family in brain development and neurological disease. Commun. Integr. Biol. 2013, 6, e26839. [Google Scholar] [CrossRef] [Green Version]
- Katrancha, S.M.; Wu, Y.; Zhu, M.; Eipper, B.; Koleske, A.J.; Mains, R.E. Neurodevelopmental disease-associated de novo mutations and rare sequence variants affect TRIO GDP/GTP exchange factor activity. Hum. Mol. Genet. 2017, 26, 4728–4740. [Google Scholar] [CrossRef]
- Su, Y.-T.; Lau, S.-F.; Ip, J.P.K.; Cheung, K.; Cheung, T.H.T.; Fu, A.K.Y.; Ip, N.Y. α2-Chimaerin is essential for neural stem cell homeostasis in mouse adult neurogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 13651–13660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadokoro, K.; Hashimoto, R.; Tatsumi, M.; Kosuga, A.; Kamijima, K.; Kunugi, H. The Gem interacting protein (GMIP) gene is associated with major depressive disorder. Neurogenetics 2005, 6, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chai, A.; Wang, L.; Ma, Y.; Wu, Z.; Yu, H.; Mei, L.; Lu, L.; Zhang, C.; Yue, W.; et al. Synaptic P-Rex1 signaling regulates hippocampal long-term depression and autism-like social behavior. Proc. Natl. Acad. Sci. USA 2015, 112, E6964–E6972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, R.; Yu, C.-E.; Yu, J.; Munson, J.; Chen, D.; Hua, W.; Estes, A.; Cortes, F.; De La Barra, F.; Yu, N.; et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 2002, 80, 129–134. [Google Scholar] [CrossRef]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Abe, M.; Yamazaki, M.; Sakimura, K.; Yamada, K.; Hoshino, M. Heterozygous Disruption of Autism susceptibility candidate 2 Causes Impaired Emotional Control and Cognitive Memory. PLoS ONE 2015, 10, e0145979. [Google Scholar] [CrossRef]
- Bedogni, F.; Hodge, R.D.; Nelson, B.R.; Frederick, E.; Shiba, N.; Daza, R.A.; Hevner, R.F. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr. Patterns 2010, 10, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Taya, S.; Hashimoto, R.; Hayashi, T.; Abe, M.; Yamazaki, M.; Nakao, K.; et al. Cytoskeletal Regulation by AUTS2 in Neuronal Migration and Neuritogenesis. Cell Rep. 2014, 9, 2166–2179. [Google Scholar] [CrossRef] [Green Version]
- Maestrini, E.; Pagnamenta, A.T.; Lamb, J.; Bacchelli, E.; Sykes, N.H.; Sousa, I.; Toma, C.; Barnby, G.; Butler, H.; Winchester, L.; et al. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L–DOCK4 gene region in autism susceptibility. Mol. Psychiatry 2009, 15, 954–968. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Peng, Y.; Wan, J.; Tang, G.; Chen, Y.; Tang, J.; Ye, W.-C.; Ip, N.Y.; Shi, L. The Atypical Guanine Nucleotide Exchange Factor Dock4 Regulates Neurite Differentiation through Modulation of Rac1 GTPase and Actin Dynamics. J. Biol. Chem. 2013, 288, 20034–20045. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Peng, Y.; Wang, L.; Sun, X.; Wang, X.; Liang, C.; Yang, X.; Li, S.; Xu, J.; Ye, W.-C.; et al. Autism-like social deficit generated by Dock4 deficiency is rescued by restoration of Rac1 activity and NMDA receptor function. Mol. Psychiatry 2019, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Guo, H.; Xiong, B.; Stessman, H.A.; Wu, H.; Coe, B.P.; Turner, T.N.; Liu, Y.; Zhao, W.; Hoekzema, K.; et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 2016, 7, 13316. [Google Scholar] [CrossRef] [PubMed]
- Okabe, T.; Nakamura, T.; Nishimura, Y.N.; Kohu, K.; Ohwada, S.; Morishita, Y.; Akiyama, T. RICS, a Novel GTPase-activating Protein for Cdc42 and Rac1, Is Involved in the β-Catenin-N-cadherin andN-Methyl-d-aspartate Receptor Signaling. J. Biol. Chem. 2003, 278, 9920–9927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Ma, H.; Bossy-Wetzel, E.; Lipton, S.A.; Zhang, Z.; Feng, G.-S. GC-GAP, a Rho Family GTPase-activating Protein That Interacts with Signaling Adapters Gab1 and Gab2. J. Biol. Chem. 2003, 278, 34641–34653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Arima-Yoshida, F.; Sakaue, F.; Nasu-Nishimura, Y.; Takeda, Y.; Matsuura, K.; Akshoomoff, N.; Mattson, S.N.; Grossfeld, P.D.; Manabe, T.; et al. PX-RICS-deficient mice mimic autism spectrum disorder in Jacobsen syndrome through impaired GABA A receptor trafficking. Nat. Commun. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Sakaue, F.; Nasu-Nishimura, Y.; Takeda, Y.; Matsuura, K.; Akiyama, T. The Autism-Related Protein PX-RICS Mediates GABAergic Synaptic Plasticity in Hippocampal Neurons and Emotional Learning in Mice. EBioMedicine 2018, 34, 189–200. [Google Scholar] [CrossRef]
- Datta, D.; Arion, M.; Corradi, J.; Lewis, D.A. Altered expression of CDC42 signaling pathway components in cortical layer 3 pyramidal cells in schizophrenia. Biol. Psychiatry 2015, 78, 775–785. [Google Scholar] [CrossRef] [Green Version]
- Davidkova, G.; McCullumsmith, R.E.; Meador-Woodruff, J.H. Expression of ARHGEF11 mRNA in schizophrenic thalamus. Ann. N. Y. Acad. Sci. 2003, 1003, 375–377. [Google Scholar] [CrossRef]
- Hashimoto, R.; Yoshida, M.; Ozaki, N.; Yamanouchi, Y.; Iwata, N.; Suzuki, T.; Kitajima, T.; Tatsumi, M.; Kamijima, K.; Kunugi, H. A missense polymorphism (H204R) of a Rho GTPase-activating protein, the chimerin 2 gene, is associated with schizophrenia in men. Schizophr. Res. 2005, 73, 383–385. [Google Scholar] [CrossRef]
- Zanni, G.; Saillour, Y.; Nagara, M.; Billuart, P.; Castelnau, L.; Moraine, C.; Faivre, L.; Bertini, E.; Dürr, A.; Guichet, A.; et al. Oligophrenin 1 mutations frequently cause X-linked mental retardation with cerebellar hypoplasia. Neurology 2005, 65, 1364–1369. [Google Scholar] [CrossRef]
- Piton, A.; Gauthier, J.; Hamdan, F.F.; Lafrenière, R.G.; Yang, Y.; Henrion, E.; Laurent, S.; Noreau, A.; Thibodeau, P.; Karemera, L.; et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol. Psychiatry 2010, 16, 867–880. [Google Scholar] [CrossRef]
- Barresi, S.; Tomaselli, S.; Athanasiadis, A.; Galeano, F.; Locatelli, F.; Bertini, E.; Zanni, G.; Gallo, A. Oligophrenin-1 (OPHN1), a Gene Involved in X-Linked Intellectual Disability, Undergoes RNA Editing and Alternative Splicing during Human Brain Development. PLoS ONE 2014, 9, e91351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khelfaoui, M.; Denis, C.; Van Galen, E.; De Bock, F.; Schmitt, A.; Houbron, C.; Morice, E.; Giros, B.; Ramakers, G.; Fagni, L.; et al. Loss of X-Linked Mental Retardation Gene Oligophrenin1 in Mice Impairs Spatial Memory and Leads to Ventricular Enlargement and Dendritic Spine Immaturity. J. Neurosci. 2007, 27, 9439–9450. [Google Scholar] [CrossRef] [PubMed]
- Meziane, H.; Khelfaoui, M.; Morello, N.; Hiba, B.; Calcagno, E.; Reibel-Foisset, S.; Selloum, M.; Chelly, J.; Humeau, Y.; Riet, F.; et al. Fasudil treatment in adult reverses behavioural changes and brain ventricular enlargement in Oligophrenin-1 mouse model of intellectual disability. Hum. Mol. Genet. 2016, 25, 2314–2323. [Google Scholar] [CrossRef] [PubMed]
- Khelfaoui, M.; Gambino, F.; Houbaert, X.; Ragazzon, B.; Müller, C.; Carta, M.; Lanore, F.; Srikumar, B.N.; Gastrein, P.; Lepleux, M.; et al. Lack of the presynaptic RhoGAP protein oligophrenin1 leads to cognitive disabilities through dysregulation of the cAMP/PKA signalling pathway. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, T.; Bathoorn, A.; Ahmadian, M.R.; Collard, J.G. Identification and characterization of hPEM-2, a guanine nucleotide exchange factor specific for Cdc42. J. Biol. Chem. 1999, 274, 33587–33593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalscheuer, V.M.; Musante, L.; Fang, C.; Hoffmann, K.; Fuchs, C.; Carta, E.; Deas, E.; Venkateswarlu, K.; Menzel, C.; Ullmann, R.; et al. A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum. Mutat. 2009, 30, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulos, T.; Korte, M.; Eulenburg, V.; Kubota, H.; Retiounskaia, M.; Harvey, R.J.; Harvey, K.; O’Sullivan, G.; Laube, B.; Hülsmann, S.; et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 2007, 26, 3888–3899. [Google Scholar] [CrossRef]
- Carlson, B.R.; Lloyd, K.E.; Kruszewski, A.; Kim, I.-H.; Rodriguiz, R.M.; Heindel, C.; Faytell, M.; Dudek, S.M.; Wetsel, W.C.; Soderling, S.H. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 2011, 31, 2447–2460. [Google Scholar] [CrossRef] [Green Version]
- Waltereit, R.; Leimer, U.; Halbach, O.V.B.U.; Panke, J.; Hölter, S.M.; Garrett, L.; Wittig, K.; Schneider, M.; Schmitt, C.; Calzada-Wack, J.; et al. Srgap3−/− mice present a neurodevelopmental disorder with schizophrenia-related intermediate phenotypes. FASEB J. 2012, 26, 4418–4428. [Google Scholar] [CrossRef]
- Valdez, C.M.; Murphy, G.G.; Beg, A.A. The Rac-GAP alpha2-chimaerin regulates hippocampal dendrite and spine morphogenesis. Mol. Cell. Neurosci. 2016, 75, 14–26. [Google Scholar] [CrossRef] [Green Version]
- Luuk, H.; Kõks, S.; Plaas, M.; Hannibal, J.; Rehfeld, J.F.; Vasar, E. Distribution of Wfs1 protein in the central nervous system of the mouse and its relation to clinical symptoms of the Wolfram syndrome. J. Comp. Neurol. 2008, 509, 642–660. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-M.; Huang, J.-P.; Eipper, B.A.; Mains, R.E. Expression of Trio, a member of the Dbl family of Rho GEFs in the developing rat brain. J. Comp. Neurol. 2005, 482, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Nuss, P. Anxiety disorders and GABA neurotransmission: A disturbance of modulation. Neuropsychiatr. Dis. Treat. 2015, 11, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, T.; Fogaça, M.V.; Deyama, S.; Li, X.-Y.; Fukumoto, K.; Duman, R.S. BDNF release and signaling are required for the antidepressant actions of GLYX-13. Mol. Psychiatry 2017, 23, 2007–2017. [Google Scholar] [CrossRef]
- Patrício, P.; Mateus-Pinheiro, A.; Irmler, M.; Alves, N.D.; Machado-Santos, A.R.; Morais, M.; Correia, J.S.; Korostynski, M.; Piechota, M.; Stoffel, R.; et al. Differential and Converging Molecular Mechanisms of Antidepressants’ Action in the Hippocampal Dentate Gyrus. Neuropsychopharmacology 2014, 40, 338–349. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-L.; Chen, J.-J.; Han, F.; Pan, C.; Zhuang, T.-T.; Cai, Y.; Lü, Y.-P. Novel antidepressant effects of Paeonol alleviate neuronal injury with concomitant alterations in BDNF, Rac1 and RhoA levels in chronic unpredictable mild stress rats. Psychopharmacology 2018, 235, 2177–2191. [Google Scholar] [CrossRef]
- Redolfi, N.; Galla, L.; Maset, A.; Murru, L.; Savoia, E.; Zamparo, I.; Grittia, A.; Billuart, P.; Passafaro, M.; Lodovichi, C. Oligophrenin-1 regulates number, morphol ogy and synaptic properties of adult-born inhibitory interneurons in the olfactory bulb. Hum. Mol. Genet. 2016, 25, 5198–5211. [Google Scholar] [CrossRef] [Green Version]
- Reddy, J.M.; Raut, N.G.R.; Seifert, J.L.; Hynds, D.L. Regulation of Small GTPase Prenylation in the Nervous System. Mol. Neurobiol. 2020, 1–12. [Google Scholar] [CrossRef]
- Kim, G.; McKee, A.E.; Ning, Y.-M.; Hazarika, M.; Theoret, M.; Johnson, J.R.; Xu, Q.C.; Tang, S.; Sridhara, R.; Jiang, X.; et al. FDA Approval Summary: Vemurafenib for Treatment of Unresectable or Metastatic Melanoma with the BRAFV600E Mutation. Clin. Cancer Res. 2014, 20, 4994–5000. [Google Scholar] [CrossRef] [Green Version]




| GTPase | Study Model | Study Findings | Ref. |
|---|---|---|---|
| RhoA | Neuronal culture | Inhibits dendritic growth and dynamics; constitutive overexpression leads to spine loss | [48] |
| Rac1 | Neuronal culture | Promotes dendritic growth and dynamics | [48] |
| Hippocampal slice culture | Overexpression of dominant-negative Rac1 reduces spine density | [49] | |
| Conditional knock-out (alpha CaMKII promoter, deletion in the postnatal hippocampus) | Reduction in Rac1 activity and spine density, impairment in learning and memory (using Rac1 inhibitors) | [50,51] | |
| Cdc42 | Neuronal culture | Promotes dendritic growth and dynamics | [48] |
| Conditional knock-out (alpha CaMKII promoter, deletion in postnatal forebrain) | Spine density of hippocampal CA1 reduced, remote memory recall was impaired | [52] |
| GTPase | BLA Expression | CEA Expression | Ref. |
|---|---|---|---|
| RhoA | + (in all parts of the amygdala) | [67] | |
| Rac1 | + | not reported | [74,77] |
| Cdc42 | + * | - | |
| Kalirin | + | CEl and CEm | [78] |
| Rich2 | + | [79] | |
| BCR | + * | [80] | |
| Dock9 | + * | [80] | |
| Trio | + * | [80] | |
| Wfs1 | + | [81] | |
| Auts2 | + * | [80] | |
| Dock4 | + * | [80] | |
| RICS | + | [82] | |
| RAPGEF6 | + | [83] | |
| Oligophrenin 1 | ubiquitous (whole brain) | [84] | |
| MEGAP | + | [85] | |
| GTPase/Regulator | Function in Amygdala | Association with Disease | Ref. |
|---|---|---|---|
| Rac1 | Extinction of contextual fear | [71] | |
| Reconsolidation of auditory fear memory | [73] | ||
| Post-training activation is required for both long-term and short-term auditory fear memory. | [74] | ||
| Photoactivation of Rac1 in LA impairs fear memory. | [75] | ||
| Alpha2- chimaerin (RhoGAP) | Abnormal contextual fear learning | [145] | |
| ArhGAP4 | Odor stimulus avoidance | [65] | |
| MEGAP (ArhGAP14) | Loss of MEGAP disrupts learning and memory | ASD and ID | [143] |
| ArhGAP32 | Knock-out showed abnormal fear learning memory and ASD-like behaviors | ASD | [129,130] |
| Oligophrenin 1 (RhoGAP) | Deletion leads to a deficit in fear memory extinction and alteration in the cAMP/PKA pathway | ASD, ID, and Cerebellar Hypoplasia | [134,135,139] |
| RhoGEF KIAA0377 | Odor stimulus avoidance | [65] | |
| Wfs1 | Modulation of anxiety and fear | Bipolar disease (Wolfram disease) | [146] |
| TRIO (RhoGEF) | Neural Development | Bipolar disease | [115,147] |
| Collybistin (ArhGEF9) | Deletion leads to loss of inhibitory neurons | ID, Anxiety, Epilepsy | [131] |
| ArhGEF10 | Knock-out increased norepinephrine and serotonin levels in the amygdala | ASD | [118] |
| RAPGEF6 | Deletion leads to reduced fear learning and less neuronal activation | SCZ | [83] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarowar, T.; Grabrucker, A.M. Rho GTPases in the Amygdala—A Switch for Fears? Cells 2020, 9, 1972. https://doi.org/10.3390/cells9091972
Sarowar T, Grabrucker AM. Rho GTPases in the Amygdala—A Switch for Fears? Cells. 2020; 9(9):1972. https://doi.org/10.3390/cells9091972
Chicago/Turabian StyleSarowar, Tasnuva, and Andreas M. Grabrucker. 2020. "Rho GTPases in the Amygdala—A Switch for Fears?" Cells 9, no. 9: 1972. https://doi.org/10.3390/cells9091972