An Integrative Study Showing the Adaptation to Sub-Optimal Growth Conditions of Natural Populations of Arabidopsis thaliana: A Focus on Cell Wall Changes
Abstract
:1. Introduction
2. Materials and Methods
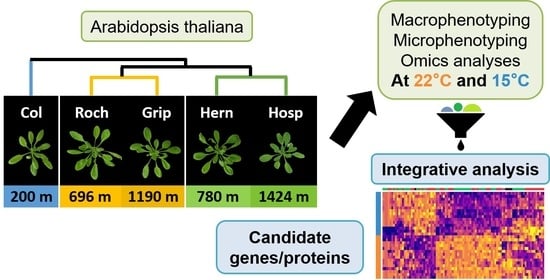
2.1. Plant Material
2.2. Macrophenotyping
2.3. Histological Staining of CWs
2.4. Extraction of Proteins from Purified CWs
2.5. Sequential CW Polysaccharides Extraction and Identification
2.6. Identification of Proteins by LC-MS/MS
2.7. RNA Sequencing
2.8. Bioinformatics Annotation of Proteins and Quantification
2.9. Statistical Analysis and Data Integration
3. Results
3.1. The Morphological Phenotypes Are Temperature- and Organ-Dependent
3.2. The CW Composition of Aerial Organs Varies Depending upon Temperature Growth Conditions
3.3. The CW Proteome Varies at Sub-Optimal Growth Temperature
3.4. Regulation of Gene Expression Depends on Both Growth Temperatures and Populations
3.5. An Integrative Study Highlights the Different Responses of the Rosettes and Floral Stems According to the Growth Temperature
3.6. The Hosp and the Hern Populations Exhibit Particular Phenotypic Responses Regarding Rosette and Floral Stem Development
4. Discussion
4.1. Temperature Has an Important Impact on Plant Development and on the CW
4.2. The Response to Growth Temperature is Organ-Specific
4.3. The Hern and Hosp Populations Exhibit Particularities in Their Responses to Growth Temperature
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CW | cell wall |
| CWP | cell wall proteins |
| LTP | lipid transfer protein |
| PME | pectin methylesterase |
References
- Bisbis, M.B.; Gruda, N.; Blanke, M. Potential impacts of climate change on vegetable production and product quality–A review. J. Clean. Prod. 2018, 170, 1602–1620. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Hanumantharao, B.; Nair, R.M.; Vara Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.M.; Varshney, R.K.; et al. Food legumes and rising temperatures: Effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso-Blanco, C.; Mendez-Vigo, B.; Koornneef, M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int. J. Dev. Biol. 2005, 49, 717–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, P.B.; Wright, I.J.; Cavender-Bares, J.; Craine, J.M.; Oleksyn, J.; Westoby, M.; Walters, M.B. The evolution of plant functional variation: Traits, spectra, and strategies. Int. J. Plant Sci. 2003, 164, S143–S164. [Google Scholar] [CrossRef]
- Körner, C. The use of “altitude” in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- Byars, S.G.; Papst, W.; Hoffmann, A.A. Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution 2007, 61, 2925–2941. [Google Scholar] [CrossRef]
- Gonzalo-Turpin, H.; Hazard, L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J. Ecol. 2009, 97, 742–751. [Google Scholar] [CrossRef]
- Brousseau, L.; Postolache, D.; Lascoux, M.; Drouzas, A.D.; Källman, T.; Leonarduzzi, C.; Liepelt, S.; Piotti, A.; Popescu, F.; Roschanski, A.M.; et al. Local adaptation in European firs assessed through extensive sampling across altitudinal gradients in southern Europe. PLoS ONE 2016, 11, e0158216. [Google Scholar] [CrossRef]
- Sultan, S.E. Promising directions in plant phenotypic plasticity. Perspect. Plant Ecol. Evol. Syst. 2004, 6, 227–233. [Google Scholar] [CrossRef] [Green Version]
- Auge, G.A.; Leverett, L.D.; Edwards, B.R.; Donohue, K. Adjusting phenotypes via within- and across-generational plasticity. New Phytol. 2017, 216, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Hamann, E.; Scheepens, J.F.; Kesselring, H.; Armbruster, G.F.J.; Stöcklin, J. High intraspecific phenotypic variation, but little evidence for local adaptation in Geum reptans populations in the Central Swiss Alps. Alp. Bot. 2017, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Völler, E.; Bossdorf, O.; Prati, D.; Auge, H. Evolutionary responses to land use in eight common grassland plants. J. Ecol. 2017, 105, 1290–1297. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Weyand, A.; Rudmann-Maurer, K.; Stöcklin, J. Adaptation of Poa alpina to altitude and land use in the Swiss Alps. Alp. Bot. 2011, 121, 91–105. [Google Scholar] [CrossRef] [Green Version]
- Trontin, C.; Tisné, S.; Bach, L.; Loudet, O. What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr. Opin. Plant Biol. 2011, 14, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Delker, C.; Quint, M. Expression level polymorphisms: Heritable traits shaping natural variation. Trends Plant Sci. 2011, 16, 481–488. [Google Scholar] [CrossRef]
- Duruflé, H.; Hervé, V.; Ranocha, P.; Balliau, T.; Zivy, M.; Chourré, J.; San Clemente, H.; Burlat, V.; Albenne, C.; Déjean, S.; et al. Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions: An integrative study. Plant Sci. 2017, 263. [Google Scholar] [CrossRef]
- Braidwood, L.; Breuer, C.; Sugimoto, K. My body is a cage: Mechanisms and modulation of plant cell growth. New Phytol. 2014, 201, 388–402. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Baldwin, L.; Domon, J.M.; Klimek, J.K.; Fournet, F.; Sellier, H.; Gillet, F.; Pelloux, J.; Lejeune-Hénaut, I.; Carpita, N.C.; Rayon, R. Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 1994, 104, 37–47. [Google Scholar] [CrossRef]
- Ingram, G.; Nawrath, C. The roles of the cuticle in plant development: Organ adhesions and beyond. J. Exp. Bot. 2017, 68, 5307–5321. [Google Scholar] [CrossRef]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasidharan, R.; Voesenek, L.A.C.J.; Pierik, R. Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit. Rev. Plant Sci. 2011, 30, 548–562. [Google Scholar] [CrossRef]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that “cut and paste” plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef]
- Montesinos, A.; Tonsor, S.J.; Alonso-Blanco, C.; Picó, F.X. Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 2009, 4, e7213. [Google Scholar] [CrossRef] [Green Version]
- Pico, F.X. Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. J. Ecol. 2012, 100, 1009–1018. [Google Scholar] [CrossRef] [Green Version]
- Botto, J.F. Plasticity to simulated shade is associated with altitude in structured populations of Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 1321–1332. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Widmer, A.; Karrenberg, S. The roles of genetic drift and natural selection in quantitative trait divergence along an altitudinal gradient in Arabidopsis thaliana. Heredity 2014, 114, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Suter, L.; Widmer, A. Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS ONE 2013, 8, e80819. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Kattge, J.; Cornelissen, J.H.C.; Wright, I.J.; Lavorel, S.; Dray, S.; Reu, B.; Kleyer, M.; Wirth, C.; Prentice, I.C.; et al. The global spectrum of plant form and function. Nature 2016, 529, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Singh, S.; Mishra, P.; Singh, A.; Tripathi, A.M.; Jena, S.N.; Roy, S. Genetic diversity and population structure of Arabidopsis thaliana along an altitudinal gradient. AoB Plants 2015, 8, plv145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Günther, T.; Lampei, C.; Barilar, I.; Schmid, K.J. Genomic and phenotypic differentiation of Arabidopsis thaliana along altitudinal gradients in the North Italian Alps. Mol. Ecol. 2016, 25, 3574–3592. [Google Scholar] [CrossRef] [PubMed]
- Lampei, C.; Wunder, J.; Wilhalm, T.; Schmid, K.J. Microclimate predicts frost-hardiness of alpine Arabidopsis thaliana populations better than altitude because the microclimate effect increases with altitude. Ecol. Evol. 2019, 9, 13017–13029. [Google Scholar] [CrossRef] [Green Version]
- Scheepens, J.; Deng, Y.; Bosdorff, O. Phenotypic plasticity in response to temperature fluctuations is genetically variable, and relates to climatic variability of origin, in Arabidopsis thaliana. AoB Plants 2018, 10, ply043. [Google Scholar] [CrossRef]
- Ibañez, C.; Poeschl, Y.; Peterson, T.; Bellstädt, J.; Denk, K.; Gogol-Döring, A.; Quint, M.; Delker, C. Ambient temperature and genotype differentially affect developmental and phenotypic plasticity in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 114. [Google Scholar] [CrossRef] [Green Version]
- Duruflé, H.; Ranocha, P.; Mbadinga Mbadinga, D.L.; Déjean, S.; Bonhomme, M.; San Clemente, H.; Hinoux, V.; Sáez-Vásquez, J.; Reichheld, J.-P.; Escaravage, N.; et al. Phenotypic trait variation as a response to altitude-related constraints in Arabidopsis populations. Front. Plant Sci. 2019, 10, 430. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Duruflé, H.; Selmani, M.; Ranocha, P.; Jamet, E.; Dunand, C.; Déjean, S. A powerful framework for an integrative study with heterogeneous omics data: From univariate statistics to multi-block analysis. Brief. Bioinform. 2020, bbaa166. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; Albenne, C.; Jamet, E.; Dunand, C. Phenotyping and cell wall polysaccharide composition dataset of five arabidopsis ecotypes grown at optimal or sub-optimal temperatures. Data Brief 2019, 25, 104318. [Google Scholar] [CrossRef] [PubMed]
- Francoz, E.; Ranocha, P.; Pernot, C.; Ru, A.L.; Pacquit, V.; Dunand, C.; Burlat, V. Complementarity of medium-throughput in situ RNA hybridization and tissue-specific transcriptomics: Case study of Arabidopsis seed development kinetics. Sci. Rep. 2016, 6, 24644. [Google Scholar] [CrossRef] [PubMed]
- Feiz, L.; Irshad, M.; Pont-Lezica, R.F.; Canut, H.; Jamet, E. Evaluation of cell wall preparations for proteomics: A new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2006, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Houben, K.; Jolie, R.P.; Fraeye, I.; Van Loey, A.M.; Hendrickx, M.E. Comparative study of the cell wall composition of broccoli, carrot, and tomato: Structural characterization of the extractable pectins and hemicelluloses. Carbohydr. Res. 2011, 346, 1105–1111. [Google Scholar] [CrossRef]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef]
- Langella, O.; Valot, B.; Balliau, T.; Blein-Nicolas, M.; Bonhomme, L.; Zivy, M. X ! TandemPipeline: A tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res. 2016, 16, 494–503. [Google Scholar] [CrossRef]
- Duruflé, H.; Ranocha, P.; Balliau, T.; Dunand, C.; Jamet, E. Transcriptomic and cell wall proteomic datasets of rosettes and floral stems from five Arabidopsis thaliana ecotypes grown at optimal or sub-optimal temperature. Data Brief 2019, 27, 104581. [Google Scholar] [CrossRef]
- San Clemente, H.; Pont-Lezica, R.; Jamet, E. Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins: Plant cell wall proteomics as a test case. Bioinform. Biol. Insights 2009, 3, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valot, B.; Langella, O.; Nano, E.; Zivy, M. MassChroQ: A versatile tool for mass spectrometry quantification. Proteomics 2011, 11, 3572–3577. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; Clemente, H.S.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 2017, 17, 1600449. [Google Scholar] [CrossRef] [PubMed]
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Masson, P.H.; Somerville, C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648. [Google Scholar] [CrossRef] [Green Version]
- Califar, B.; Sng, N.J.; Zupanska, A.; Paul, A.L.; Ferl, R.J. Root Skewing-Associated Genes Impact the Spaceflight Response of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 239. [Google Scholar] [CrossRef] [Green Version]
- Domon, J.M.; Baldwin, L.; Acket, S.; Caudeville, E.; Arnoult, S.; Zub, H.; Gillet, F.; Lejeune-Hénaut, I.; Brancourt-Hulmel, M.; Pelloux, J.; et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 2013, 85, 51–61. [Google Scholar] [CrossRef]
- Parrotta, L.; Faleri, C.; Guerriero, G.; Cai, G. Cold stress affects cell wall deposition and growth pattern in tobacco pollen tubes. Plant Sci. 2019, 283, 329–342. [Google Scholar] [CrossRef]
- Sharma, N.; Cram, D.; Huebert, T.; Zhou, N.; Parkin, I.A. Exploiting the wild crucifer Thlaspi arvense to identify conserved and novel genes expressed during a plant’s response to cold stress. Plant Mol. Biol. 2007, 63, 171–184. [Google Scholar] [CrossRef]
- Oono, Y.; Seki, M.; Satou, M.; Iida, K.; Akiyama, K.; Sakurai, T.; Fujita, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct. Integr. Genom. 2006, 6, 212–234. [Google Scholar] [CrossRef]
- Imaizumi, C.; Tomatsu, H.; Kitazawa, K.; Yoshimi, Y.; Shibano, S.; Kikuchi, K.; Yamaguchi, M.; Kaneko, S.; Tsumuraya, Y.; Kotake, T. Heterologous expression and characterization of an Arabidopsis β-l-arabinopyranosidase and α-d-galactosidases acting on β-l-arabinopyranosyl residues. J. Exp. Bot. 2017, 68, 4651–4661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, W.; Jia, Y. The serine carboxypeptidase-like gene SCPL41 negatively regulates membrane lipid metabolism in Arabidopsis thaliana. Plants 2020, 9, 696. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Xiong, W.; Ye, T.; Wu, Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J. Exp. Bot. 2012, 63, 2579–2593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef]
- Lee, S.C.; Kim, S.H.; An, S.H.; Yi, S.Y.; Hwang, B.K. Identification and functional expression of the pepper pathogen-induced gene, CAPIP2, involved in disease resistance and drought and salt stress tolerance. Plant Mol. Biol. 2006, 62, 151–164. [Google Scholar] [CrossRef]
- Majid, M.U.; Awan, M.F.; Fatima, K.; Tahir, M.S.; Ali, Q.; Rashid, B.; Rao, A.Q.; Nasir, I.A.; Husnain, T. Genetic resources of chili pepper (Capsicum annuum L.) against Phytophthora capsici and their induction through various biotic and abiotic factors. Cytol. Genet. 2017, 51, 296–304. [Google Scholar] [CrossRef]
- Suh, M.C.; Samuels, A.L.; Jetter, R.; Kunst, L.; Pollard, M.; Ohlrogge, J.; Beisson, F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005, 139, 1649–1665. [Google Scholar] [CrossRef] [Green Version]
- Jacq, A.; Pernot, C.; Martinez, Y.; Domergue, F.; Payré, B.; Jamet, E.; Burlat, V.; Pacquit, V. The Arabidopsis Lipid Transfer Protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Sci. 2017, 8, 263. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.E.; Li, Y.; Labbe, A.; Guevara, D.; Nuin, P.; Whitty, B.; Diaz, C.; Golding, G.B.; Gray, G.R.; Weretilnyk, E.A.; et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef] [Green Version]
- Lyou, S.H.; Park, H.J.; Jung, C.; Sohn, H.B.; Lee, G.; Kim, C.H.; Kim, M.; Choi, Y.D.; Cheong, J.J. The Arabidopsis AtLEC gene encoding a lectin-like protein is up-regulated by multiple stimuli including developmental signal, wounding, jasmonate, ethylene, and chitin elicitor. Mol. Cells 2009, 27, 75–81. [Google Scholar] [CrossRef]
- Catinot, J.; Huang, J.B.; Huang, P.Y.; Tseng, M.Y.; Chen, Y.L.; Gu, S.Y.; Lo, W.S.; Wang, L.C.; Chen, Y.R.; Zimmerli, L. ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. 2015, 38, 2721–2734. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.Y.; Zhong, G.V.; Burns, J.K. Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 2003, 53, 117–131. [Google Scholar] [CrossRef]
- Schaller, A. A cut above the rest: The regulatory function of plant proteases. Planta 2004, 220, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Fanourakis, D.; Nikoloudakis, N.; Pappi, P.; Markakis, E.; Doupis, G.; Charova, S.N.; Delis, C.; Tsaniklidis, G. The role of proteases in determining stomatal development and tuning pore aperture: A review. Plants 2020, 9, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Lee, S.B.; Kim, H.J.; Min, M.K.; Hwang, I.; Suh, M.C. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1391–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, T.; Liu, R.; Wang, W.; An, L.; Chen, T.; Liu, G.; Zhao, Z. Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology 2011, 63, 111–117. [Google Scholar] [CrossRef] [PubMed]







| Accession Name | Altitude (MASL) 1 | Genetics Cluster 2 | Climate PC1 3 |
|---|---|---|---|
| Col | 200 4 | III | 3 |
| Roch | 696 | I | 0.7 |
| Grip | 1190 | I | −1.4 |
| Hern | 780 | II | 0.7 |
| Hosp | 1424 | II | −1.6 |
| AGI Code | Functional Class 1 | Putative Function | Proteins 2 | Transcripts 2 | 22 °C 2 | 15 °C 2 |
|---|---|---|---|---|---|---|
| AT2G45180 | LM | homologous to non-specific lipid transfer protein | + | + | ||
| AT1G54010 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT5G15350 | OR | early nodulin (AtEN22, AtENODL17), blue copper binding protein | + | + | ||
| AT4G37800 | PAC | GH16 (endoxyloglucan transferase) (AtXTH7) | + | + | ||
| AT3G10740 | PAC | GH51 (α-arabinofuranosidase) | + | + | ||
| AT2G45470 | S | fasciclin-like arabinogalactan protein (AtFLA8) | + | + | ||
| AT3G18050 | UF | expressed protein | + | + | ||
| AT1G53070 | ID | lectin (legume lectin domain) | + | + | ||
| AT1G33590 | ID | expressed protein (LRR domains) | + | + | ||
| AT3G12610 | ID | expressed protein (LRR domains) | + | + | ||
| AT3G62820 | ID | plant invertase/pectin methylesterase inhibitor (PMEI) | + | + | ||
| AT5G59320 | LM | non-specific lipid transfer protein (AtLTP1.2, AtLTP3) | + | + | ||
| AT5G59310 | LM | non-specific lipid transfer protein (AtLTP1.11, AtLTP4) | + | + | ||
| AT3G53980 | LM | non-specific lipid transfer protein (AtLTPd7) | + | + | ||
| AT1G41830 | OR | multicopper oxidase (AtSKS6, homologous to SKU5) | + | + | ||
| AT2G23000 | P | Ser carboxypeptidase (AtSCPL10) | + | + | ||
| AT3G18490 | P | Asp protease (pepsin family) (ASPG1) | + | + | ||
| AT1G29050 | PAC | homologous to A. thaliana PMR5 (carbohydrate acylation) | + | + | ||
| AT1G21250 | S | receptor kinase (AtWAK1, WALL-ASSOCIATED KINASE 1) | + | + | ||
| AT1G16850 | UF | expressed protein | + | + | ||
| AT1G33811 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT2G37640 | PAC | α-expansin (ATHEXP ALPHA 1.9) (AtEXPA3) | + | + | ||
| AT3G22060 | UF | expressed protein | + | + | ||
| AT5G14450 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT2G27190 | M | purple acid phosphatase (AtPAP12) | + | + | ||
| AT1G24450 | M | ribonuclease III | + | + | ||
| AT1G07080 | OR | thiol reductase (GILT family) | + | + | ||
| AT4G21960 | OR | class III peroxidase (AtPrx42) | + | + | ||
| AT4G36195 | P | Pro-Xaa carboxypeptidase (Peptidase family S28.A26, MEROPS) | + | + | ||
| AT5G65760 | P | Pro-Xaa carboxypeptidase (Peptidase family S28.A02, MEROPS) | + | + | ||
| AT4G34980 | P | Ser protease (AtSBT1.6) | + | + | ||
| AT3G10410 | P | Ser carboxypeptidase (AtSCPL49) | + | + | ||
| AT4G30810 | P | Ser carboxypeptidase (AtSCPL29) | + | + | ||
| AT3G26380 | PAC | GH27 (α-galactosidase/melibiase, APSE) | + | + | ||
| AT5G08370 | PAC | GH27 (α-galactosidase/melibiase, AGAL1) | + | + | ||
| AT5G08380 | PAC | GH27 (α-galactosidase/melibiase, AGAL2) | + | + | ||
| AT5G07830 | PAC | GH79 (endo-β-glucuronidase/heparanase) | + | + | ||
| AT3G05910 | PAC | CE13 (pectin acylesterase - PAE) (AtPAE12) | + | + | ||
| AT4G21620 | UF | expressed protein | + | + | ||
| AT5G39570 | UF | expressed protein | + | + |
| AGI Code 1 | Functional Class 2 | Putative Function | Proteins 3 | Transcripts 3 | 22 °C 3 | 15 °C 3 |
|---|---|---|---|---|---|---|
| AT2G10940 | LM | homologous to non-specific lipid transfer protein | + | + | ||
| AT5G21100 | OR | multicopper oxidase | + | + | ||
| AT5G15350 | OR | early nodulin (AtEN22, AtENODL17), blue copper binding protein | + | + | ||
| AT1G47128 | P | Cys protease (papain family) (Peptidase family C01.064) (RD21A) | + | + | ||
| AT4G21640 | P | Ser protease (AtSBT3.15) | + | + | ||
| AT3G10450 | P | Ser carboxypeptidase (AtSCPL7) | + | + | ||
| AT5G19740 | P | peptidase M28 (peptidase family M28.A02, MEROPS) | + | + | ||
| AT1G26560 | PAC | GH1 (β-glucosidase) (AtBGLU40) | + | + | ||
| AT2G06850 | PAC | GH16 (endoxyloglucan transferase) (At-XTH4) | + | + | ||
| AT1G10640 | PAC | GH28 (polygalacturonase) | + | + | ||
| AT3G02880 | S | leucine-rich repeat receptor protein kinase (LRR III subfamily) | + | + | ||
| AT3G22060 | UF | expressed protein (Gnk2-homologous domain) | + | + | ||
| AT5G62210 | LM | expressed protein (PLAT/LH2 domain) | + | + | ||
| AT1G31550 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT1G29670 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT1G18250 | M | thaumatin (PR5, ATLP1) | + | + | ||
| AT5G51950 | OR | expressed protein (GMC oxido-reductase domain) | + | + | ||
| AT4G31840 | OR | early nodulin (AtEN13, AtENODL15), blue copper binding protein | + | + | ||
| AT4G01700 | PAC | GH19 | + | + | ||
| AT4G23820 | PAC | GH28 (polygalacturonase) | + | + | ||
| AT5G06860 | ID | PGIP1 (LRR domains) | + | + | ||
| AT3G13990 | OR | multicopper oxidase (AtSKS11, homologous to SKU5) | + | + | ||
| AT5G20230 | OR | stellacyanin AtSTC1, BCB (blue copper binding protein) | + | + | ||
| AT1G71695 | OR | class III peroxidase (AtPrx12) | + | + | ||
| AT2G39850 | P | Ser protease (AtSBT4.1) | + | + | ||
| AT4G21585 | LM | phospholipase C/P1 nuclease | + | + | ||
| AT2G38540 | LM | non-specific lipid-transfer protein (AtLTP1.5, AtLTP1) | + | + | ||
| AT1G27950 | LM | non-specific lipid transfer protein (AtLTPg4, LTPG1) | + | + | ||
| AT5G62210 | LM | expressed protein (lipase/lipooxygenase domain, PLAT/LH2) | + | + | ||
| AT2G04570 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT1G41830 | OR | multicopper oxidase (AtSKS6, homologous to SKU5) | + | + | ||
| AT4G12880 | OR | early nodulin (AtEN20, AtENODL19), blue copper binding protein | + | + | ||
| AT2G33530 | P | Ser carboxypeptidase (AtSCPL46) | + | + | ||
| AT5G45280 | PAC | CE13 (pectin acylesterase - PAE) (AtPAE11) | + | + | ||
| AT4G38400 | PAC | expansin-like A (ATHEXP BETA 2.2) (AtEXLA2) | + | + | ||
| AT5G55730 | S | fasciclin-like arabinogalactan protein (AtFLA1) | + | + | ||
| AT2G04780 | S | fasciclin-like arabinogalactan protein (AtFLA7) | + | + | ||
| AT3G06035 | UF | expressed protein | + | + | ||
| AT4G00860 | UF | expressed protein (DUF1138) | + | + | ||
| AT3G05730 | UF | expressed protein | + | + |
| AGI Code 1 | Functional Class 2 | Putative Function | Proteins 3 | Transcripts 3 |
|---|---|---|---|---|
| AT1G78850 | ID | lectin (curculin-like) | + | |
| AT3G04720 | ID | expressed protein (Barwin domain, defense protein) | + | |
| AT1G66970 | LM | glycerophosphoryl diester phosphodiesterase (GDPDL1, GDPL3) | + | |
| AT1G55260 | LM | non-specific lipid transfer protein (AtLTPg6) | + | |
| AT2G14610 | M | PR1/Cys-rich secretory protein (SCP) | + | |
| AT1G55210 | M | dirigent protein (AtDIR20) | + | |
| AT1G78680 | P | peptidase C26 (peptidase family C26.003, MEROPS) (GGH2) | + | |
| AT5G08370 | PAC | GH27 (α-galactosidase/melibiase) | + | |
| AT3G16850 | PAC | GH28 (polygalacturonase) | + | |
| AT2G18660 | PAC | expansin-like (AtEXR3) | + | |
| AT5G38980 | UF | expressed protein | + | |
| AT3G28220 | UF | expressed protein (MATH domain) | + | |
| AT5G48540 | UF | expressed protein | + | |
| AT4G29240 | ID | expressed protein (LRR domains) | + | |
| AT4G12390 | ID | plant invertase/pectin methylesterase inhibitor (PMEI-like) | + | |
| AT1G49740 | LM | expressed protein (phospholipase C domain) | + | |
| AT5G03610 | LM | lipase acylhydrolase (GDSL family) | + | |
| AT3G08770 | LM | non-specific lipid transfer protein (AtLTP1.6, AtLTP6) | + | |
| AT3G14920 | PAC | peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidase A | + | |
| AT5G38980 | UF | expressed protein | + | |
| AT4G28100 | UF | expressed protein | + | |
| AT2G47010 | UF | expressed protein | + |
| AGI Code 1 | Functional Class 2 | Putative Function | Proteins 3 | Transcripts 3 | Hosp 3 | Hern 3 |
|---|---|---|---|---|---|---|
| AT1G66970 | LM | glycerophosphoryl diester phosphodiesterase (GDPDL1, GDPL3) | + | + | ||
| AT1G27950 | LM | non-specific lipid transfer protein (AtLTPg4, LTPG1) | + | + | ||
| AT3G32980 | OR | class III peroxidase (AtPrx32) (ATP16A) | + | + | ||
| AT2G12480 | P | Ser carboxypeptidase (AtSCPL43) | + | + | ||
| AT3G02740 | P | Asp protease (peptidase family A01.A23, MEROPS) | + | + | ||
| AT4G31140 | PAC | GH17 (β-1,3-glucosidase) | + | + | ||
| AT5G60950 | S | AtCOBL5 | + | + | ||
| AT5G19240 | UF | expressed protein | + | + | ||
| AT3G15356 | ID | lectin (legume lectin domain) | + | + | ||
| AT3G16530 | ID | lectin (legume lectin domain) | + | + | ||
| AT3G04720 | ID | expressed protein (PR4, Barwin domain, defense protein) | + | + | ||
| AT2G26010 | M | homologous to gamma thionin (defensin) | + | + | ||
| AT4G16260 | PAC | GH17 (β-1,3-glucosidase) | + | + | ||
| AT1G29050 | PAC | homologous to A. thaliana PMR5 (carbohydrate acylation) | + | + | ||
| AT1G78820 | ID | lectin (curculin-like) | + | + | ||
| AT2G42800 | ID | expressed protein (LRR domains) | + | + | ||
| AT2G17120 | ID | expressed protein (LysM domain) | + | + | ||
| AT5G03610 | LM | lipase acylhydrolase (GDSL family) | + | + | ||
| AT4G39640 | M | gamma glutamyltranspeptidase (GGT2) | + | + | ||
| AT4G02330 | PAC | CE8 (AtPME41) | + | + | ||
| AT5G09760 | PAC | CE8 (AtPME51) | + | + | ||
| AT1G78060 | PAC | GH3 (β-xylosidase) (AtBXL7) | + | + | ||
| AT5G42100 | PAC | GH17 (β -1,3-glucosidase) | + | + | ||
| AT4G34480 | PAC | GH17 (β -1,3-glucosidase) | + | + | ||
| AT4G01700 | PAC | GH19 | + | + | ||
| AT3G02880 | S | recepteur kinase (LRR III subfamily) | + | + | ||
| AT5G19240 | UF | expressed protein | + | + | ||
| AT5G48540 | UF | expressed protein | + | + | ||
| AT3G44100 | LM | expressed protein (MD-2-related lipid-recognition)domain | + | - | ||
| AT1G31550 | LM | lipase acylhydrolase (GDSL family) | + | - | ||
| AT1G01300 | P | Asp protease (peptidase family A01.A05, MEROPS) | + | - | ||
| AT3G61820 | P | Asp protease (peptidase family A01.A13, MEROPS) | + | - | ||
| AT3G12700 | P | Asp protease (peptidase family A01.A30, MEROPS) | + | - | ||
| AT3G52500 | P | Asp protease (peptidase family A01.A47, MEROPS) | + | - | ||
| AT5G19740 | P | peptidase M28 (peptidase family M28.A02, MEROPS) | + | - | ||
| AT2G22970 | P | Ser carboxypeptidase (AtSCPL11) | + | - | ||
| AT3G02110 | P | Ser carboxypeptidase (AtSCPL25) | + | - | ||
| AT5G23210 | P | Ser carboxypeptidase (AtSCPL34) | + | - | ||
| AT1G28110 | P | Ser carboxypeptidase (AtSCPL45) | + | - | ||
| AT5G51750 | P | Ser protease (AtSBT1.3) | + | - | ||
| AT4G00230 | P | Ser protease (AtSBT4.14, XSP1) | + | - | ||
| AT5G25980 | PAC | GH1 (AtBGLU37) | + | - | ||
| AT1G70710 | PAC | GH9 (AtCEL1) | + | - | ||
| AT4G02290 | PAC | GH9 (endo-1,3(4)-β-glucanase) | + | - | ||
| AT4G23820 | PAC | GH28 (polygalacturonase) | + | - | ||
| AT4G33220 | PAC | CE8 (pectin methylesterase – PME) (AtPME44) | + | - | ||
| AT5G23870 | PAC | CE13 (pectin acylesterase - PAE) (AtPAE9) | + | - | ||
| AT1G04680 | PAC | PL1 (pectate lyase) (AtPLL26) | + | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duruflé, H.; Ranocha, P.; Balliau, T.; Zivy, M.; Albenne, C.; Burlat, V.; Déjean, S.; Jamet, E.; Dunand, C. An Integrative Study Showing the Adaptation to Sub-Optimal Growth Conditions of Natural Populations of Arabidopsis thaliana: A Focus on Cell Wall Changes. Cells 2020, 9, 2249. https://doi.org/10.3390/cells9102249
Duruflé H, Ranocha P, Balliau T, Zivy M, Albenne C, Burlat V, Déjean S, Jamet E, Dunand C. An Integrative Study Showing the Adaptation to Sub-Optimal Growth Conditions of Natural Populations of Arabidopsis thaliana: A Focus on Cell Wall Changes. Cells. 2020; 9(10):2249. https://doi.org/10.3390/cells9102249
Chicago/Turabian StyleDuruflé, Harold, Philippe Ranocha, Thierry Balliau, Michel Zivy, Cécile Albenne, Vincent Burlat, Sébastien Déjean, Elisabeth Jamet, and Christophe Dunand. 2020. "An Integrative Study Showing the Adaptation to Sub-Optimal Growth Conditions of Natural Populations of Arabidopsis thaliana: A Focus on Cell Wall Changes" Cells 9, no. 10: 2249. https://doi.org/10.3390/cells9102249