Alternative Splicing in Heat Shock Protein Transcripts as a Mechanism of Cell Adaptation in Trichophyton rubrum
Abstract
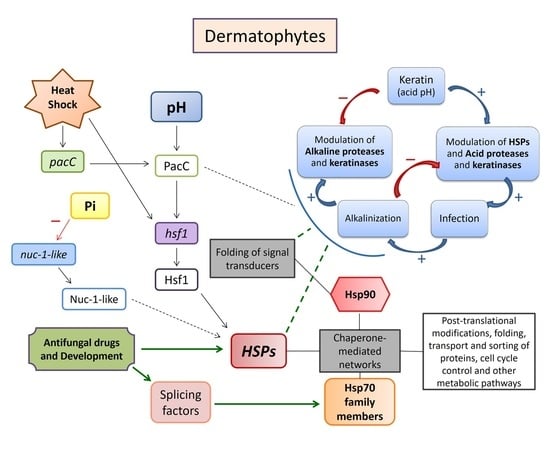
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Alternative Splicing Assay in Response to Drugs
2.3. Co-Culture Assay with Human Keratinocytes
2.4. Keratinolytic Assay
2.5. RNA Extraction and cDNA Synthesis
2.6. In Silico Characterization of HSP Isoforms
2.7. Gene Expression Analysis
3. Results
3.1. Alternative Splicing Assay
3.2. HSPs Expression Profile in An Infection-Like Scenario
3.3. Evaluation of the Keratinolytic Potential of T. rubrum under Hsp90 Inhibition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M.J. Heat shock proteins. J. Biol. Chem. 1990, 265, 12111–12114. [Google Scholar] [PubMed]
- Csermely, P.; Yahara, I. Heat shock proteins. In Molecular Pathomechanics and New Trends in Drug Research; Keri, G., Toth, I., Eds.; CRC Press: London, UK, 2003; pp. 67–75. [Google Scholar]
- Rikhvanov, E.G.; Romanova, N.V.; Chernoff, Y.O. Chaperone effects on prion and nonprion aggregates. Prion 2007, 1, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Burnie, J.P.; Carter, T.L.; Hodgetts, S.J.; Matthews, R.C. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 2006, 30, 53–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.; Thakur, R.; Shankar, J. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnol. Res. Int. 2015, 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Jacob, T.R.; Peres, N.T.A.; Martins, M.P.; Lang, E.A.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. Heat Shock Protein 90 (Hsp90) as a Molecular Target for the Development of Novel Drugs Against the Dermatophyte Trichophyton rubrum. Front. Microbiol. 2015, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Jacob, T.R.; Sanches, P.R.; Peres, N.T.A.; Lang, E.A.S.; Martins, M.P.; Rossi, A. Heat shock proteins in dermatophytes: Current advances and perspectives. Curr. Genom. 2016, 17, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Peres, N.T.A.; Rossi, A. Antifungal resistance mechanisms in dermatophytes. Mycopathologia 2008, 166, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Persinoti, G.F.; Peres, N.T.A.; Rossi, A. Role of pH in the pathogenesis of dermatophytoses. Mycoses 2011, 55, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Peres, N.T.A.; Maranhão, F.C.; Rossi, A.; Martinez-Rossi, N.M. Dermatophytes: Host-pathogen interaction and antifungal resistance. An. Bras. Dermatol. 2010, 85, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Rodwell, G.E.; Bayles, C.L.; Towersey, L.; Aly, R. The prevalence of dermatophyte infection in patients infected with human immunodeficiency virus. Int. J. Dermatol. 2008, 47, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Faure-Cognet, O.; Fricker-Hidalgo, H.; Pelloux, H.; Leccia, M.T. Superficial Fungal Infections in a French Teaching Hospital in Grenoble Area: Retrospective Study on 5470 Samples from 2001 to 2011. Mycopathologia 2015, 181, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Mezzari, A. Frequency of dermatophytes in the metropolitan area of Porto Alegre, RS, Brazil. Rev. Inst. Med. Trop. Sao Paulo 1998, 40, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Nenoff, P.; Kruger, C.; Ginter-Hanselmayer, G.; Tietz, H.J. Mycology—An update. Part 1: Dermatomycoses: Causative agents, epidemiology and pathogenesis. J. Dtsch. Dermatol. Ges. 2014, 12, 188–209. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, C.; Bouchara, J.P.; Mignon, B. Updates on the epidemiology of dermatophyte infections. Mycopathologia 2008, 166, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Leal, J.; Squina, F.M.; Freitas, J.S.; Silva, E.M.; Ono, C.J.; Martinez-Rossi, N.M.; Rossi, A. A splice variant of the Neurospora crassa hex-1 transcript, which encodes the major protein of the Woronin body, is modulated by extracellular phosphate and pH changes. FEBS Lett. 2009, 583, 180–184. [Google Scholar] [CrossRef]
- Mendes, N.S.; Silva, P.M.; Silva-Rocha, R.; Martinez-Rossi, N.M.; Rossi, A. Pre-mRNA splicing is modulated by antifungal drugs in the filamentous fungus Neurospora crassa. FEBS Open Bio 2016, 6, 358–368. [Google Scholar] [CrossRef]
- Trevisan, G.L.; Oliveira, E.H.; Peres, N.T.A.; Cruz, A.H.; Martinez-Rossi, N.M.; Rossi, A. Transcription of Aspergillus nidulans pacC is modulated by alternative RNA splicing of palB. FEBS Lett. 2011, 585, 3442–3445. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Oliveira, F.B.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. The prp4 kinase gene and related spliceosome factor genes in Trichophyton rubrum respond to nutrients and antifungals. J. Med. Microbiol. 2019, 68, 591–599. [Google Scholar] [CrossRef]
- Grutzmann, K.; Szafranski, K.; Pohl, M.; Voigt, K.; Petzold, A.; Schuster, S. Fungal Alternative Splicing is Associated with Multicellular Complexity and Virulence: A Genome-Wide Multi-Species Study. DNA Res. 2014, 21, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Kempken, F. Alternative splicing in ascomycetes. Appl. Microbiol. Biotechnol. 2013, 97, 4235–4241. [Google Scholar] [CrossRef]
- Jin, L.; Li, G.; Yu, D.; Huang, W.; Cheng, C.; Liao, S.; Wu, Q.; Zhang, Y. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae. BMC Genom. 2017, 18, 130. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Voigt, K.; Kammer, P.; Brunke, S.; Schuster, S.; Linde, J. Comparative Study on Alternative Splicing in Human Fungal Pathogens Suggests Its Involvement During Host Invasion. Front. Microbiol. 2018, 9, 13. [Google Scholar] [CrossRef]
- Zhao, C.; Waalwijk, C.; de Wit, P.J.; Tang, D.; van der Lee, T. RNA-Seq analysis reveals new gene models and alternative splicing in the fungal pathogen Fusarium graminearum. BMC Genom. 2013, 14, 16. [Google Scholar] [CrossRef]
- Ast, G. How did alternative splicing evolve? Nat. Rev. Genet. 2004, 5, 773–782. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell 2006, 126, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Peres, N.T.A.; Silva, L.G.; Santos Rda, S.; Jacob, T.R.; Persinoti, G.F.; Rocha, L.B.; Falcao, J.P.; Rossi, A.; Martinez-Rossi, N.M. In vitro and ex vivo infection models help assess the molecular aspects of the interaction of Trichophyton rubrum with the host milieu. Med. Mycol. 2016, 54, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Komoto, T.T.; Bitencourt, T.A.; Silva, G.; Beleboni, R.O.; Marins, M.; Fachin, A.L. Gene Expression Response of Trichophyton rubrum during Coculture on Keratinocytes Exposed to Antifungal Agents. Evid. Based. Complement. Alternat. Med. 2015, 2015, 180535. [Google Scholar] [CrossRef] [PubMed]
- Gradisar, H.; Friedrich, J.; Krizaj, I.; Jerala, R. Similarities and specificities of fungal keratinolytic proteases: Comparison of keratinases of Paecilomyces marquandii and Doratomyces microsporus to some known proteases. Appl. Environ. Microbiol. 2005, 71, 3420–3426. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Macedo, C.; Franco, M.E.; Assis, A.F.; Komoto, T.T.; Stehling, E.G.; Beleboni, R.O.; Malavazi, I.; Marins, M.; Fachin, A.L. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genom. 2016, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Mendes, N.S.; Bitencourt, T.A.; Sanches, P.R.; Silva-Rocha, R.; Martinez-Rossi, N.M.; Rossi, A. Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Sci. Rep. 2018, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2008, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jacob, T.R.; Peres, N.T.A.; Persinoti, G.F.; Silva, L.G.; Mazucato, M.; Rossi, A.; Martinez-Rossi, N.M. rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 2012, 50, 368–377. [Google Scholar] [CrossRef]
- Leach, M.D.; Klipp, E.; Cowen, L.E.; Brown, A.J. Fungal Hsp90: A biological transistor that tunes cellular outputs to thermal inputs. Nat. Rev. Microbiol. 2012, 10, 693–704. [Google Scholar] [CrossRef]
- Lindquist, S. Protein folding sculpting evolutionary change. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 103–108. [Google Scholar] [CrossRef]
- Peccarelli, M.; Kebaara, B.W. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot. Cell 2014, 13, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Longman, D.; Caceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell 2016, 59, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Dang, Y.; Zhou, M.; Li, L.; Yu, C.H.; Fu, J.; Chen, S.; Liu, Y. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. USA 2016, 113, E6117–E6125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, J.P.B. The evolution and diversity of the nonsense-mediated mRNA decay pathway. F1000Research 2018, 7, 11. [Google Scholar] [CrossRef]
- Zhou, Z.; Dang, Y.; Zhou, M.; Yuan, H.; Liu, Y. Codon usage biases co-evolve with transcription termination machinery to suppress premature cleavage and polyadenylation. Elife 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.V.; Bortolossi, J.C.; Sanches, P.R.; Mendes, N.S.; Martinez-Rossi, N.M.; Rossi, A. STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum. Int. J. Mol. Sci. 2018, 19, 3654. [Google Scholar] [CrossRef] [PubMed]
- Zaugg, C.; Monod, M.; Weber, J.; Harshman, K.; Pradervand, S.; Thomas, J.; Bueno, M.; Giddey, K.; Staib, P. Gene expression profiling in the human pathogenic dermatophyte Trichophyton rubrum during growth on proteins. Eukaryot. Cell 2009, 8, 241–250. [Google Scholar] [CrossRef]
- Martinez-Rossi, N.M.; Peres, N.T.; Rossi, A. Pathogenesis of Dermatophytosis: Sensing the Host Tissue. Mycopathologia 2017, 182, 215–227. [Google Scholar] [CrossRef]






| Gene ID | Putative Annotation | Sequence 5′-3′ | Concentration (nM) | Size (bp) |
|---|---|---|---|---|
| TERG_03206 RT-PCR | hsp7-like | F: ATGCTCGCCTCAAGAATCTG R: ATCAATACCGATGACCTGGC | - | 241/153 |
| TERG_03206 qPCR | hsp7-like | F: CCAGGTCATCGGTATTGATCTT R: CGGATGGAGTGGTTCTTGTT | 100 | 110 |
| TERG_03206 qPCR | hsp-7 like intron-1 | F: GTAAGTAGCAAGCTGCAAGG R: CTAGACGGTTAAATGTCAGTCAAA | 100 | 88 |
| TERG_01883 qPCR | hsp75-like | F: GAAAGAGGTTGCCGAGACTAAG R: CTCTGGTTGTCGTTGAAGTAGG | 300 | 84 |
| TERG_01659 qPCR | hsp30 | F: AAAGTTCGATGTGAGGGAGTG R: GAGTGTTTGAGGATCGGTGAA | 100 | 106 |
| TERG_04141 qPCR | hsp60 | F: GATTGCCCAGGTTGCTACTAT R: AGTGATGACACCTTCCTTTCC | 100 | 100 |
| TERG_06963 qPCR | hsp90 | F: GGTCAACAACCTCGGTACTATT R: GACACCGAACTGTCCAATCA | 300 | 100 |
| TERG_07658 qPCR | hsp70-like | F: TGAGATGGTCGGTGGATGTA R: GGTTTAGGGTGAAGGAGAGTTG | 300 | 89 |
| TERG_07949 qPCR | hsp104 | F: TGTCGAGAAGGACGAAGAATAAC R: GGAACATCGCCTCTGACTATC | 300 | 106 |
| TERG_12507 qPCR | hsp104-like | F: GCTGATGGATGATGGTCGTATC R: GTTGGGCGGTTCAGGTATT | 100 | 111 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves-da-Rocha, J.; Bitencourt, T.A.; de Oliveira, V.M.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. Alternative Splicing in Heat Shock Protein Transcripts as a Mechanism of Cell Adaptation in Trichophyton rubrum. Cells 2019, 8, 1206. https://doi.org/10.3390/cells8101206
Neves-da-Rocha J, Bitencourt TA, de Oliveira VM, Sanches PR, Rossi A, Martinez-Rossi NM. Alternative Splicing in Heat Shock Protein Transcripts as a Mechanism of Cell Adaptation in Trichophyton rubrum. Cells. 2019; 8(10):1206. https://doi.org/10.3390/cells8101206
Chicago/Turabian StyleNeves-da-Rocha, João, Tamires A. Bitencourt, Vanderci M. de Oliveira, Pablo R. Sanches, Antonio Rossi, and Nilce M. Martinez-Rossi. 2019. "Alternative Splicing in Heat Shock Protein Transcripts as a Mechanism of Cell Adaptation in Trichophyton rubrum" Cells 8, no. 10: 1206. https://doi.org/10.3390/cells8101206