Understanding Bartonella-Associated Infective Endocarditis: Examining Heart Valve and Vegetation Appearance and the Role of Neutrophilic Leukocytes
Abstract
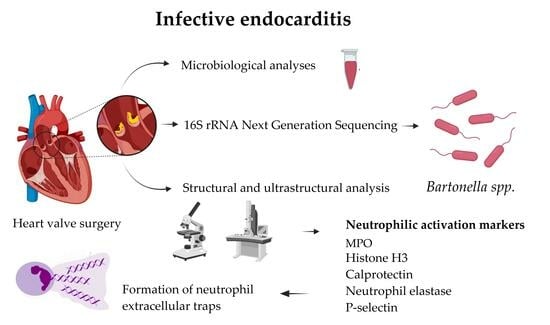
:1. Introduction
2. Materials and Methods
2.1. Patients’ Characteristics and General Clinical Data
2.2. Methods of Microbiological Investigation
2.2.1. Microbial DNA Extraction for 16S rRNA NGS Analyses
2.2.2. 16S rRNA Gene V3-V4 Amplification and Illumina MiSeq Sequencing
2.2.3. 16S Sequence Analysis
2.3. Histopathological and Histochemical Investigation of the Heart Valve Alteration in IE
2.4. Immunohistochemical Investigation of the Heart Valve Leaflets and Vegetations in IE
2.5. Transmission Electron Microscopy of the Heart Valve Leaflets and Vegetations in IE
2.6. Scanning Electron Microscopy of the Heart Valve Leaflets and Vegetations in IE
2.7. Statistical Data Analysis
3. Results
3.1. Patients’ Characteristics, Laboratory Indices, and Clinical Outcomes in Non-Bartonella spp.- and Bartonella spp.-Caused IE
3.2. 16S rRNA NGS Testing Results for IE
3.3. Histopathological and Histochemical Assessment of the Heart Valve Alteration and Vegetation in IE
3.4. Immunohistochemical Assessment of the Expression of Markers of Neutrophil Activation Contributing to NETosis in the Heart Valve Leaflets and Vegetations in IE
3.5. Multivariate Analysis of Immunohistochemically Obtained Expressions of Neutrophilic Leukocyte Activation Markers
3.6. Ultrastructural Analysis of the Heart Valve Leaflets and Vegetations in IE
3.7. Hierarchical Clustering Used for the Exploration of Data Similarities and Visualization Using Alluvial Plotting
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Talha, K.M.; Baddour, L.M.; Thornhill, M.H.; Arshad, V.; Tariq, W.; Tleyjeh, I.M.; Scott, C.G.; Hyun, M.H.; Bailey, K.R.; Anavekar, N.S.; et al. Escalating incidence of infective endocarditis in Europe in the 21st century. Open Hear. 2021, 8, e001846. [Google Scholar] [CrossRef] [PubMed]
- Lisi, M.; Flamigni, F.; Russo, M.; Cameli, M.; Mandoli, G.E.; Pastore, M.C.; Mele, D.; Campo, G.; Henein, M.Y.; Rubolli, A. Incidence and mortality of infective endocarditis in the last decade: A single center study. J. Cardiovasc. Med. 2023, 24, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rajani, R.; Klein, J.L. Infective endocarditis: A contemporary update. Clin. Med. 2020, 20, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Sexton, D.J.; Spelman, D. Current best practices and guidelines. Assessment and management of complications in infective endocarditis. Cardiol. Clin. 2003, 21, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Chirillo, F. New approach to managing infective endocarditis. Trends Cardiovasc. Med. 2021, 31, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Friedrich, C.; Saad, M.; Frank, D.; Salem, M.; Puehler, T.; Schoettler, J.; Schoeneich, F.; Cremer, J.; Haneya, A. Active Infective Native and Prosthetic Valve Endocarditis: Short- and Long-Term Outcomes of Patients after Surgical Treatment. J. Clin. Med. 2021, 10, 1868. [Google Scholar] [CrossRef]
- Luaces, M.; Vilacosta, I.; Fernández, C.; Sarriá, C.; San Román, J.A.; Graupner, C.; Núñez-Gil, I.J. Vegetation size at diagnosis in infective endocarditis: Influencing factors and prognostic implications. Int. J. Cardiol. 2009, 137, 76–78. [Google Scholar] [CrossRef]
- Leitman, M.; Dreznik, Y.; Tyomkin, V.; Fuchs, T.; Krakover, R.; Vered, Z. Vegetation size in patients with infective endocarditis. Eur. Heart J. Cardiovasc. Imaging 2012, 13, 330–338. [Google Scholar] [CrossRef]
- Vikram, H.R. The long and short of vegetations in infective endocarditis. Expert Rev. Anti-Infect. Ther. 2007, 5, 529–533. [Google Scholar] [CrossRef]
- Kamde, S.P.; Anjankar, A. Pathogenesis, Diagnosis, Antimicrobial Therapy, and Management of Infective Endocarditis, and Its Complications. Cureus 2022, 14, e29182. [Google Scholar] [CrossRef]
- Braï, M.A.; Hannachi, N.; El Gueddari, N.; Baudoin, J.P.; Dahmani, A.; Lepidi, H.; Habib, G.; Camoin-Jau, L. The Role of Platelets in Infective Endocarditis. Int. J. Mol. Sci. 2023, 24, 7540. [Google Scholar] [CrossRef] [PubMed]
- Luehr, M.; Bauernschmitt, N.; Peterss, S.; Li, Y.; Heyn, O.; Dashkevich, A.; Oberbach, A.; Bagaev, E.; Pichlmaier, M.A.; Juchem, G.; et al. Incidence and Surgical Outcomes of Patients with Native and Prosthetic Aortic Valve Endocarditis. Ann. Thorac. Surg. 2020, 110, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Meyers, S.; Crescente, M.; Verhamme, P.; Martinod, K. Staphylococcus aureus and Neutrophil Extracellular Traps: The Master Manipulator Meets Its Match in Immunothrombosis. Arter. Thromb. Vasc. Biol. 2022, 42, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Nappi, F.; Martuscelli, G.; Bellomo, F.; Singh, S.S.A.; Moon, M.R. Infective Endocarditis in High-Income Countries. Metabolites 2022, 12, 682. [Google Scholar] [CrossRef] [PubMed]
- Haddad, S.F.; DeSimone, D.C.; Chesdachai, S.; Gerberi, D.J.; Baddour, L.M. Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review. Antibiotics 2022, 11, 1798. [Google Scholar] [CrossRef]
- Suardi, L.R.; de Alarcon, A.; Garcia, M.V.; Ciezar, A.P.; Hidalgo Tenorio, C.; Martinez-Marcos, F.J.; Concejo-Martinez, E.; De la Torre Lima, J.; Vinuesa Garcia, D.; Luque Marquez, R.; et al. Blood culture-negative infective endocarditis: A worse outcome? Results from a large multicentre retrospective Spanish cohort study. Infect. Dis. 2021, 53, 755–763. [Google Scholar] [CrossRef]
- Scheer, C.S.; Fuchs, C.; Gründling, M.; Vollmer, M.; Bast, J.; Bohnert, J.A.; Zimmermann, K.; Hahnenkamp, K.; Rehberg, S.; Kuhn, S.O. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin. Microbiol. Infect. 2019, 25, 326–331. [Google Scholar] [CrossRef]
- Okaro, U.; Addisu, A.; Casanas, B.; Anderson, B. Bartonella Species, an Emerging Cause of Blood-Culture-Negative Endocarditis. Clin. Microbiol. Rev. 2017, 30, 709–746. [Google Scholar] [CrossRef]
- Liesenborghs, L.; Meyers, S.; Vanassche, T.; Verhamme, P. Coagulation: At the heart of infective endocarditis. J. Thromb. Haemost. 2020, 18, 995–1008. [Google Scholar] [CrossRef]
- Hidalgo, A.; Libby, P.; Soehnlein, O.; Aramburu, I.V.; Papayannopoulos, V.; Silvestre-Roig, C. Neutrophil extracellular traps: From physiology to pathology. Cardiovasc. Res. 2022, 118, 2737–2753. [Google Scholar] [CrossRef]
- Tanaka, Y.; Yamanaka, N.; Koyano, I.; Hasunuma, I.; Kobayashi, T.; Kikuyama, S.; Iwamuro, S. Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action. Antibiotics 2022, 11, 1240. [Google Scholar] [CrossRef]
- Meyers, S.; Lox, M.; Kraisin, S.; Liesenborghs, L.; Martens, C.P.; Frederix, L.; Van Bruggen, S.; Crescente, M.; Missiakas, D.; Baatsen, P.; et al. Neutrophils Protect Against Staphylococcus aureus Endocarditis Progression Independent of Extracellular Trap Release. Arter. Thromb. Vasc. Biol. 2023, 43, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.R.; Witten, J.C.; Tan, C.D.; Rodriguez, E.R.; Blackstone, E.H.; Pettersson, G.B.; Seifert, D.E.; Willard, B.B.; Apte, S.S. Proteomics identifies a convergent innate response to infective endocarditis and extensive proteolysis in vegetation components. J. Clin. Investig. 2020, 5, e135317. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lu, W.; Ning, Y.; Liu, J. Reverse effects of Streptococcus mutans physiological states on neutrophil extracellular traps formation as a strategy to escape neutrophil killing. Front. Cell. Infect. Microbiol. 2022, 12, 1023457. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.J.; Radic, M. Neutrophil Extracellular Traps: Double-Edged Swords of Innate Immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Jenne, C.N.; Wong, C.H.; Zemp, F.J.; McDonald, B.; Rahman, M.M.; Forsyth, P.A.; McFadden, G.; Kubes, P. Neutrophils Recruited to Sites of Infection Protect from Virus Challenge by Releasing Neutrophil Extracellular Traps. Cell Host Microbe 2013, 13, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J.; Fadrosh, D.W.; Ma, B.; Gajer, P.; et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Garvey, W. Silver Impregnation Techniques to Identify Spirochetes and Other Bacteria. J. Histotechnol. 1996, 19, 203–209. [Google Scholar] [CrossRef]
- McCormick, J.K.; Tripp, T.J.; Dunny, G.M.; Schlievert, P.M. Formation of Vegetations during Infective Endocarditis Excludes Binding of Bacterial-Specific Host Antibodies to Enterococcus faecalis. J. Infect. Dis. 2002, 185, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Chavakis, T.; Bierhaus, A.; Al-Fakhri, N.; Schneider, D.; Witte, S.; Linn, T.; Nagashima, M.; Morser, J.; Arnold, B.; Preissner, K.T.; et al. The pattern recognition receptor (rage) is a counterreceptor for leukocyte integrins: A novel pathway for inflammatory cell recruitment. J. Exp. Med. 2003, 198, 1507–1515. [Google Scholar] [CrossRef]
- Shaw, S.K.; Ma, S.; Kim, M.B.; Rao, R.M.; Hartman, C.U.; Froio, R.M.; Yang, L.; Jones, T.; Liu, Y.; Nusrat, A.; et al. Coordinated Redistribution of Leukocyte LFA-1 and Endothelial Cell ICAM-1 Accompany Neutrophil Transmigration. J. Exp. Med. 2004, 200, 1571–1580. [Google Scholar] [CrossRef]
- Chavakis, T.; Preissner, K.T.; Herrmann, M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007, 28, 408–418. [Google Scholar] [CrossRef]
- Thiene, G.; Basso, C. Pathology and pathogenesis of infective endocarditis in native heart valves. Cardiovasc. Pathol. 2006, 15, 256–263. [Google Scholar] [CrossRef]
- Jensen, H.E.; Gyllensten, J.; Hofman, C.; Leifsson, P.S.; Agerholm, J.S.; Boye, M.; Aalbæk, B. Histologic and Bacteriologic Findings in Valvular Endocarditis of Slaughter-Age Pigs. J. Veter- Diagn. Investig. 2010, 22, 921–927. [Google Scholar] [CrossRef]
- Patel, S.; Richert, M.E.; White, R.; Lambing, T.; Saleeb, P. A Case of Bartonella Quintana Culture-Negative Endocarditis. Am. J. Case Rep. 2019, 20, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil Extracellular Traps Contain Calprotectin, a Cytosolic Protein Complex Involved in Host Defense against Candida albicans. PLoS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.-J.; Yeh, C.-Y.; Hsu, R.-B.; Lee, C.-M.; Shun, C.-T.; Chia, J.-S. Endocarditis Pathogen Promotes Vegetation Formation by Inducing Intravascular Neutrophil Extracellular Traps Through Activated Platelets. Circ. 2015, 131, 571–581. [Google Scholar] [CrossRef] [PubMed]
- LeGuyader, A.; Watanabe, R.; Berbé, J.; Boumediene, A.; Cogné, M.; Laskar, M. Platelet activation after aortic prosthetic valve surgery. Interact. Cardiovasc. Thorac. Surg. 2006, 5, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.W.; Rumbaut, R.E. Platelets mediate acetaminophen hepatotoxicity. Blood 2015, 126, 1738–1739. [Google Scholar] [CrossRef] [PubMed]
- Pfister, C.; Pfrommer, H.; Tatagiba, M.S.; Roser, F. Vascular endothelial growth factor signals through platelet-derived growth factor receptor β in meningiomas in vitro. Br. J. Cancer 2012, 107, 1702–1713. [Google Scholar] [CrossRef] [PubMed]
- Martos, L.; Oto, J.; Fernández-Pardo, Á.; Plana, E.; Solmoirago, M.J.; Cana, F.; Hervás, D.; Bonanad, S.; Ferrando, F.; España, F.; et al. Increase of Neutrophil Activation Markers in Venous Thrombosis—Contribution of Circulating Activated Protein C. Int. J. Mol. Sci. 2020, 21, 5651. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Berends, E.T.; Chan, R.; Schwab, E.; Roy, S.; Sen, C.K.; Torres, V.J.; Wozniak, D.J. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc. Natl. Acad. Sci. USA 2018, 115, 7416–7421. [Google Scholar] [CrossRef]
- Bassani, B.; Cucchiara, M.; Butera, A.; Kayali, O.; Chiesa, A.; Palano, M.T.; Olmeo, F.; Gallazzi, M.; Dellavia, C.P.; Mortara, L.; et al. Neutrophils’ Contribution to Periodontitis and Periodontitis-Associated Cardiovascular Diseases. Int. J. Mol. Sci. 2023, 24, 15370. [Google Scholar]
- Koh, C.C.; Gollob, K.J.; Dutra, W.O. Balancing the functions of DNA extracellular traps in intracellular parasite infections: Implications for host defense, disease pathology and therapy. Cell Death Dis. 2023, 14, 450. [Google Scholar] [CrossRef]
- Burger, P.C.; Wagner, D.D. Platelet P-selectin facilitates atherosclerotic lesion development. Blood 2003, 101, 2661–2666. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef] [PubMed]
- de Bont, C.; Pruijn, G.J.M. Citrulline is not a major determinant of autoantibody reactivity to neutrophil extracellular traps. Philos. Trans. R. Soc. B Biol. Sci. 2023, 378, 20220249. [Google Scholar] [CrossRef]
- Kao, P.H.-N.; Ch’Ng, J.-H.; Chong, K.K.L.; Stocks, C.J.; Wong, S.L.; A Kline, K. Enterococcus faecalis suppresses Staphylococcus aureus-induced NETosis and promotes bacterial survival in polymicrobial infections. FEMS Microbes 2023, 4, xtad019. [Google Scholar] [CrossRef] [PubMed]
- Neubert, E.; Meyer, D.; Rocca, F.; Günay, G.; Kwaczala-Tessmann, A.; Grandke, J.; Senger-Sander, S.; Geisler, C.; Egner, A.; Schön, M.P.; et al. Chromatin swelling drives neutrophil extracellular trap release. Nat. Commun. 2018, 9, 3767. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Rizo, V.; Martínez-Guzmán, M.A.; Iñiguez-Gutierrez, L.; García-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Manley, H.R.; Keightley, M.C.; Lieschke, G.J. The Neutrophil Nucleus: An Important Influence on Neutrophil Migration and Function. Front. Immunol. 2018, 9, 2867. [Google Scholar] [CrossRef] [PubMed]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A Novel Mechanism of Rapid Nuclear Neutrophil Extracellular Trap Formation in Response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef]
- Vorobjeva, N.V.; Chernyak, B.V. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochemistry 2020, 85, 1178–1190. [Google Scholar] [CrossRef]
- Onouchi, T.; Shiogama, K.; Mizutani, Y.; Takaki, T.; Tsutsumi, Y. Visualization of Neutrophil Extracellular Traps and Fibrin Meshwork in Human Fibrinopurulent Inflammatory Lesions: III. Correlative Light and Electron Microscopic Study. Acta Histochem. ET Cytochem. 2016, 49, 141–147. [Google Scholar] [CrossRef]
- Hannachi, N.; Lepidi, H.; Fontanini, A.; Takakura, T.; Bou-Khalil, J.; Gouriet, F.; Habib, G.; Raoult, D.; Camoin-Jau, L.; Baudoin, J.-P. A Novel Approach for Detecting Unique Variations among Infectious Bacterial Species in Endocarditic Cardiac Valve Vegetation. Cells 2020, 9, 1899. [Google Scholar] [CrossRef] [PubMed]
- Zamorano, J.; Sanz, J.; Moreno, R.; Almerıa, C.; Rodrigo, J.-L.; Samedi, M.; Herrera, D.; Aubele, A.; Mataix, L.; Serra, V.; et al. Comparison of outcome in patients with culture-negative versus culture-positive active infective endocarditis. Am. J. Cardiol. 2001, 87, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.K.; Salsano, A.; Giacobbe, D.R.; A Popescu, B.; Laroche, C.; Duval, X.; Schueler, R.; Moreo, A.; Colonna, P.; Piper, C.; et al. Outcomes of culture-negative vs. culture-positive infective endocarditis: The ESC-EORP EURO-ENDO registry. Eur. Hear. J. 2022, 43, 2770–2780. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, R.; Curtis, S.; Schilling, W.H.; James, P.R. Blood culture negative endocarditis in the modern era of 16S rRNA sequencing. Clin. Med. 2020, 20, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, O.; Liu, W.; Gan, L.; Li, X.; Ma, Q.; Hu, X.; Jian, X. Coxiella burnetii and Bartonella Endocarditis Diagnosed by Metagenomic Next-Generation Sequencing. J. Clin. Med. 2022, 11, 7150. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Fournier, P.-E.; Vandenesch, F.; Mainardi, J.-L.; Eykyn, S.J.; Nash, J.; James, E.; Benoit-Lemercier, C.; Marrie, T.J. Outcome and Treatment of Bartonella Endocarditis. Arch. Intern. Med. 2003, 163, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E.B. Bartonellosis, One Health and all creatures great and small. Veter. Dermatol. 2017, 28, 96-e21. [Google Scholar] [CrossRef]
- Bonilla, D.L.; Cole-Porse, C.; Kjemtrup, A.; Osikowicz, L.; Kosoy, M. Risk Factors for Human Lice and Bartonellosis among the Homeless, San Francisco, California, USA. Emerg. Infect. Dis. 2014, 20, 1645–1651. [Google Scholar] [CrossRef]
- Ehrlich, G.D.; Ahmed, A.; Earl, J.; Hiller, N.L.; Costerton, J.W.; Stoodley, P.; Post, J.C.; DeMeo, P.; Hu, F.Z. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol. Med Microbiol. 2010, 59, 269–279. [Google Scholar] [CrossRef]
- Raybould, J.E.; Raybould, A.L.; Morales, M.K.; Zaheer, M.; Lipkowitz, M.S.; Timpone, J.G.; Kumar, P.N. Bartonella Endocarditis and Pauci-Immune Glomerulonephritis: A Case Report and Review of the Literature. Infect. Dis. Clin. Pract. 2016, 24, 254. Available online: https://journals.lww.com/infectdis/Fulltext/2016/09000/Bartonella_Endocarditis_and_Pauci_Immune.3.aspx (accessed on 28 September 2023). [CrossRef]
- Badiee, P.; Amirghofran, A.A.; Nour, M.G.; Shafa, M.; Nemati, M.H. Incidence and Outcome of Documented Fungal Endocarditis. Int. Cardiovasc. Res. J. 2014, 8, 152–155. [Google Scholar] [PubMed]












| Name | Sequence (Illumina Adapter, Heterogeneity Spacer, 16S Region Primer) | Reference |
|---|---|---|
| 16S V3 Fw (341F) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNNNNNNCCTACGGGNGGCWGCAG | [27] |
| 16S V4 Rs (805R) | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNNNNNNGACTACHVGGGTATCTAATCC | [27] |
| Characteristic | Non-Bartonella spp. IE, N = 35 | Bartonella spp. IE, N = 11 | p Value |
|---|---|---|---|
| Age, years, mean (SD) | 55.6 (14.8) | 54.6 (9.8) | p = 0.8217 |
| Sex, male, N (%) | 26 (74.3) | 10 (9.9) | pY = 0.4551 |
| EuroScore II risk score, %, median (IQR) | 6.5 (3.6–14.3) | 6.3 (3.6–12.6) | pMW = 0.9442 |
| IE mortality risk score, %, median (IQR) | 30.2 (13.6–41.1) | 30.2 (21.5–34.9) | pMW = 0.7988 |
| Charlson Comorbidity index, points, median (IQR) | 2.5 (1–5) | 4 (3–5) | pMW = 0.1868 |
| Diabetes mellitus, N (%) | 8 (22.9) | 2 (18.2) | pY = 0.9274 |
| Alcoholism history, N (%) | 5 (14.3) | 7 (63.6) | pY = 0.0124 |
| Intravenous drug usage history, N (%) | 3 (8.6) | 0 (0.0) | pY = 0.7609 |
| Length of vegetation, mm, median (IQR) | 12.0 (0.0–18.0) | 13.0 (12.0–22.0) | pMW = 0.1644 |
| Embolism, N (%) | 8 (22.9) | 3 (27.3) | pY = 0.9158 |
| Cerebral embolism, N (%) | 7 (20.0) | 2 (18.2) | pY = 0.7618 |
| Spleen embolism, N (%) | 1 (2.9) | 1 (9.1) | pY = 0.9706 |
| Kidney embolism, N (%) | 2 (5.7) | 0 (0.0) | pY = 0.9706 |
| EF of the left ventricle, %, mean (SD) | 56.4 (7.8) | 50.6 (9.4) | p = 0.0481 |
| Right ventricle systolic pressure, mm/Hg, median (IQR) | 37.5 (30.0–56.3) | 46.5 (43.8–56.3) | pMW = 0.0627 |
| Leukocytes, count 109 mL, median (IQR) | 7.6 (5.8–9.7) | 5.2 (4.2–6.0) | pMW = 0.0038 |
| Platelets, count 109 mL, median (IQR) | 241.0 (186.0–325.0) | 174.0 (82.0–218.0) | pMW = 0.0185 |
| Red blood cells, count 109 mL, median (IQR) | 3.7 (3.2–4.4) | 3.6 (3.1–4.1) | pMW = 0.1987 |
| Hemoglobin, g/L, mean (SD) | 110.4 (22.2) | 99.1 (14.1) | p = 0.1205 |
| CRP level before surgery, mg/dL, median (IQR) | 32.8 (7.5–55.1) | 33.5 (6.6–57.0) | pMW = 0.7837 |
| CRP level 2nd day after surgery, mg/dL, median (IQR) | 185.8 (133.9–228.8) | 157.8 (94.2–220.6) | pMW = 0.2979 |
| CRP level 4th day after surgery, mg/dL, median (IQR) | 110.0 (72.7–159.3) | 118.7 (104.8–215.1) | pMW = 0.4110 |
| CRP level 6th day after surgery, mg/dL, median (IQR) | 57.6 (31.0–100.5) | 71.9 (46.3–101.3) | pMW = 0.4572 |
| Procalcitonin, ng/mL, median (IQR) | 0.12 (0.08–0.28) | 0.22 (0.07–0.38) | pMW = 0.4911 |
| BNP, ng/mL, median (IQR) | 429.8 (136.4–960.6) | 1523.0 (692.9–3708.0) | pMW = 0.0002 |
| Glucose, mkmol/L, median (IQR) | 5.4 (4.8–6.7) | 4.7 (4.3–5.6) | pMW = 0.0375 |
| Creatinine, mmol/L, median (IQR) | 77.0 (64.0–89.0) | 104.0 (78.0–283.0) | pMW = 0.0071 |
| Blood loss, first day, mL, median (IQR) | 372.5 (250.0–630.0) | 420.0 (301.3–930.0) | pMW = 0.3463 |
| Resternotomy due to acute bleeding, N (%) | 6 (17.1) | 3 (27.3) | pY = 0.7618 |
| Mechanical lung ventilation, hours, median (IQR) | 11.5 (8.0–72.0) | 14.0 (9.0–25.0) | pMW = 0.7533 |
| Usage of vasopressors, N (%) | 23 (65.7) | 8 (72.7) | pY = 0.9489 |
| Usage of beta-agonists, N (%) | 13 (37.1) | 7 (63.6) | pY = 0.2311 |
| Days intrahospital, median (IQR) | 31.0 (25.0–48.0) | 37.0 (19.0–44.0) | pMW = 0.5544 |
| Days in the ICU, median (IQR) | 3.0 (1.8–9.0) | 3.0 (2.0–8.0) | pMW = 0.9106 |
| Intrahospital death, N (%) | 4 (11.4) | 0 (0.0) | pY = 0.5755 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meidrops, K.; Groma, V.; Goldins, N.R.; Apine, L.; Skuja, S.; Svirskis, S.; Gudra, D.; Fridmanis, D.; Stradins, P. Understanding Bartonella-Associated Infective Endocarditis: Examining Heart Valve and Vegetation Appearance and the Role of Neutrophilic Leukocytes. Cells 2024, 13, 43. https://doi.org/10.3390/cells13010043
Meidrops K, Groma V, Goldins NR, Apine L, Skuja S, Svirskis S, Gudra D, Fridmanis D, Stradins P. Understanding Bartonella-Associated Infective Endocarditis: Examining Heart Valve and Vegetation Appearance and the Role of Neutrophilic Leukocytes. Cells. 2024; 13(1):43. https://doi.org/10.3390/cells13010043
Chicago/Turabian StyleMeidrops, Kristians, Valerija Groma, Niks Ricards Goldins, Lauma Apine, Sandra Skuja, Simons Svirskis, Dita Gudra, Davids Fridmanis, and Peteris Stradins. 2024. "Understanding Bartonella-Associated Infective Endocarditis: Examining Heart Valve and Vegetation Appearance and the Role of Neutrophilic Leukocytes" Cells 13, no. 1: 43. https://doi.org/10.3390/cells13010043