Combination of Genomic Landsscape and 3D Culture Functional Assays Bridges Sarcoma Phenotype to Target and Immunotherapy
Abstract
:1. Introduction
2. Genomic Landscape as Biomarker for Diagnostic and Therapeutic Purposes
3. Clinical Results of Targeted Therapy Based on Genomic Landscape
3.1. Cell Cycle and MDM2 Inhibitors
3.2. Signal Transduction Inhibitors
3.3. Current Immunotherapy and Clinical Results
4. Clinical Trials Based on Genomic Landscape and N-1 Trials
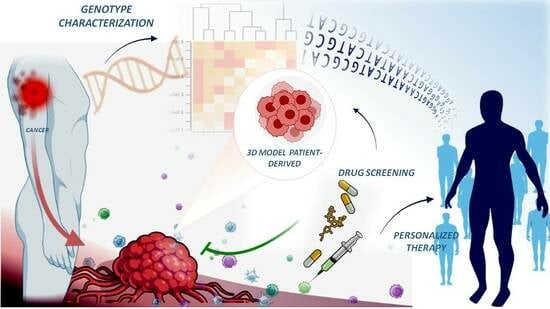
5. Individualised Patient Models
5.1. Spheroid Models
5.2. Spheroid Assays to Test Chemo- and Targeted Therapy in Preclinical and Clinical Trials
5.3. Organoid Models
5.4. Organoid Assays to Test Targeted Drugs and Chemotherapy in Preclinical and Clinical Trials
5.5. Organoid-Based Assays to Test Immunotherapy in Clinical Trials
5.6. Explants on Chip Sensor (Organ on Chip) Assays in Preclinical and Clinical Trials
6. Other Potential Evolutions
7. Prospective
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, W.J.; Doyle, L.A. Updates from the 2020 World Health Organization Classification of Soft Tissue and Bone Tumours. Histopathology 2021, 78, 644–657. [Google Scholar] [CrossRef]
- Casali, P.G.; Abecassis, N.; Aro, H.T.; Bauer, S.; Biagini, R.; Bielack, S.; Bonvalot, S.; Boukovinas, I.; Bovee, J.; Brodowicz, T.; et al. Soft tissue and visceral sarcomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29 (Suppl. S4), iv268–iv269. [Google Scholar] [CrossRef]
- de Nigris, F.; Ruosi, C.; Napoli, C. Clinical efficiency of epigenetic drugs therapy in bone malignancies. Bone 2021, 143, 115605. [Google Scholar] [CrossRef]
- Hoes, L.R.; van Berge Henegouwen, J.M.; van der Wijngaart, H.; Zeverijn, L.J.; van der Velden, D.L.; van de Haar, J.; Roepman, P.; de Leng, W.J.; Jansen, A.M.L.; van Werkhoven, E.; et al. Patients with Rare Cancers in the Drug Rediscovery Protocol (DRUP) Benefit from Genomics-Guided Treatment. Clin. Cancer Res. 2022, 28, 1402–1411. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and Trametinib in Patients With Tumors With BRAF(V600E) Mutations: Results of the NCI-MATCH Trial Subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Mechahougui, H.; Michael, M.; Friedlaender, A. Precision Oncology in Gastrointestinal Stromal Tumors. Curr. Oncol. 2023, 30, 4648–4662. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.J.; Chen, A.P.; Li, S.; McShane, L.M.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Iafrate, A.J.; Sklar, J.; et al. Molecular Landscape and Actionable Alterations in a Genomically Guided Cancer Clinical Trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J. Clin. Oncol. 2020, 38, 3883–3894. [Google Scholar] [CrossRef] [PubMed]
- Sicklick, J.K.; Kato, S.; Okamura, R.; Schwaederle, M.; Hahn, M.E.; Williams, C.B.; De, P.; Krie, A.; Piccioni, D.E.; Miller, V.A.; et al. Molecular profiling of cancer patients enables personalized combination therapy: The I-PREDICT study. Nat. Med. 2019, 25, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Palinski, W.; Monti, M.; Camerlingo, R.; Iacobucci, I.; Bocella, S.; Pinto, F.; Iannuzzi, C.; Mansueto, G.; Pignatiello, S.; Fazioli, F.; et al. Lysosome purinergic receptor P2 × 4 regulates neoangiogenesis induced by microvesicles from sarcoma patients. Cell Death Dis. 2021, 12, 797. [Google Scholar] [CrossRef] [PubMed]
- Colella, G.; Fazioli, F.; Gallo, M.; De Chiara, A.; Apice, G.; Ruosi, C.; Cimmino, A.; de Nigris, F. Sarcoma Spheroids and Organoids-Promising Tools in the Era of Personalized Medicine. Int. J. Mol. Sci. 2018, 19, 615. [Google Scholar] [CrossRef]
- Rupert, C.; Dell’ Aversana, C.; Mosca, L.; Montanaro, V.; Arcaniolo, D.; De Sio, M.; Bilancio, A.; Altucci, L.; Palinski, W.; Pili, R.; et al. Therapeutic targeting of P2 × 4 receptor and mitochondrial metabolism in clear cell renal carcinoma models. J. Exp. Clin. Cancer Res. 2023, 42, 134. [Google Scholar] [CrossRef]
- Subbiah, V. The next generation of evidence-based medicine. Nat. Med. 2023, 29, 49–58. [Google Scholar] [CrossRef]
- Turc-Carel, C.; Philip, I.; Berger, M.P.; Philip, T.; Lenoir, G.M. Chromosome study of Ewing’s sarcoma (ES) cell lines. Consistency of a reciprocal translocation t(11;22)(q24;q12). Cancer Genet. Cytogenet. 1984, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Rocques, P.J.; Crew, A.J.; Gill, S.; Shipley, J.; Chan, A.M.; Gusterson, B.A.; Cooper, C.S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 1994, 7, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Crabtree, G.R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013, 153, 71–85. [Google Scholar] [CrossRef]
- Rabbitts, T.H.; Forster, A.; Larson, R.; Nathan, P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993, 4, 175–180. [Google Scholar] [CrossRef]
- Crozat, A.; Aman, P.; Mandahl, N.; Ron, D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 1993, 363, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Seavey, C.N.; Pobbati, A.V.; Hallett, A.; Ma, S.; Reynolds, J.P.; Kanai, R.; Lamar, J.M.; Rubin, B.P. WWTR1(TAZ)-CAMTA1 gene fusion is sufficient to dysregulate YAP/TAZ signaling and drive epithelioid hemangioendothelioma tumorigenesis. Genes Dev. 2021, 35, 512–527. [Google Scholar] [CrossRef]
- Dermawan, J.K.; Rubin, B.P. The spectrum and significance of secondary (co-occurring) genetic alterations in sarcomas: The hallmarks of sarcomagenesis. J. Pathol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Nacev, B.A.; Sanchez-Vega, F.; Smith, S.A.; Antonescu, C.R.; Rosenbaum, E.; Shi, H.; Tang, C.; Socci, N.D.; Rana, S.; Gularte-Merida, R.; et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat. Commun. 2022, 13, 3405. [Google Scholar] [CrossRef]
- Movva, S.; Wen, W.; Chen, W.; Millis, S.Z.; Gatalica, Z.; Reddy, S.; von Mehren, M.; Van Tine, B.A. Multi-platform profiling of over 2000 sarcomas: Identification of biomarkers and novel therapeutic targets. Oncotarget 2015, 6, 12234–12247. [Google Scholar] [CrossRef] [PubMed]
- Barrios, C.; Castresana, J.S.; Ruiz, J.; Kreicbergs, A. Amplification of the c-myc proto-oncogene in soft tissue sarcomas. Oncology 1994, 51, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, F.; Tanaka, K.; Sakimura, R.; Matsumoto, Y.; Matsunobu, T.; Li, X.; Hanada, M.; Okada, T.; Iwamoto, Y. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. J. Biol. Chem. 2003, 278, 15105–15115. [Google Scholar] [CrossRef]
- Lee, S.B.; Kolquist, K.A.; Nichols, K.; Englert, C.; Maheswaran, S.; Ladanyi, M.; Gerald, W.L.; Haber, D.A. The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat. Genet. 1997, 17, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Ferracini, R.; Olivero, M.; Di Renzo, M.F.; Martano, M.; De Giovanni, C.; Nanni, P.; Basso, G.; Scotlandi, K.; Lollini, P.L.; Comoglio, P.M. Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene 1996, 12, 1697–1705. [Google Scholar]
- Fullenkamp, C.A.; Hall, S.L.; Jaber, O.I.; Pakalniskis, B.L.; Savage, E.C.; Savage, J.M.; Ofori-Amanfo, G.K.; Lambertz, A.M.; Ivins, S.D.; Stipp, C.S.; et al. TAZ and YAP are frequently activated oncoproteins in sarcomas. Oncotarget 2016, 7, 30094–30108. [Google Scholar] [CrossRef] [PubMed]
- Grimer, R.; Athanasou, N.; Gerrand, C.; Judson, I.; Lewis, I.; Morland, B.; Peake, D.; Seddon, B.; Whelan, J. UK Guidelines for the Management of Bone Sarcomas. Sarcoma 2010, 2010, 317462. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Mathoulin-Pelissier, S.; Cesne, A.L.; Terrier, P.; Bonvalot, S.; Collin, F.; Michels, J.J.; Blay, J.Y.; Coindre, J.M.; Bui, B. Trends in survival for patients with metastatic soft-tissue sarcoma. Cancer 2011, 117, 1049–1054. [Google Scholar] [CrossRef]
- Vanni, S.; Fausti, V.; Fonzi, E.; Liverani, C.; Miserocchi, G.; Spadazzi, C.; Cocchi, C.; Calabrese, C.; Gurrieri, L.; Riva, N.; et al. Unveiling the Genomic Basis of Chemosensitivity in Sarcomas of the Extremities: An Integrated Approach for an Unmet Clinical Need. Int. J. Mol. Sci. 2023, 24, 6926. [Google Scholar] [CrossRef]
- Foley, J.M.; Scholten, D.J., 2nd; Monks, N.R.; Cherba, D.; Monsma, D.J.; Davidson, P.; Dylewski, D.; Dykema, K.; Winn, M.E.; Steensma, M.R. Anoikis-resistant subpopulations of human osteosarcoma display significant chemoresistance and are sensitive to targeted epigenetic therapies predicted by expression profiling. J. Transl. Med. 2015, 13, 110. [Google Scholar] [CrossRef]
- Iwata, S.; Tatsumi, Y.; Yonemoto, T.; Araki, A.; Itami, M.; Kamoda, H.; Tsukanishi, T.; Hagiwara, Y.; Kinoshita, H.; Ishii, T.; et al. CDK4 overexpression is a predictive biomarker for resistance to conventional chemotherapy in patients with osteosarcoma. Oncol. Rep. 2021, 46, 135. [Google Scholar] [CrossRef]
- Kohlmeyer, J.L.; Gordon, D.J.; Tanas, M.R.; Monga, V.; Dodd, R.D.; Quelle, D.E. CDKs in Sarcoma: Mediators of Disease and Emerging Therapeutic Targets. Int. J. Mol. Sci. 2020, 21, 3018. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; An, H.J.; Kim, S.K.; Lee, S.A.; Kim, S.; Lim, S.M.; Kim, G.M.; Sohn, J.; Moon, Y.W. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int. J. Cancer 2019, 145, 1179–1188. [Google Scholar] [CrossRef]
- Hainaut, P.; Pfeifer, G.P. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb. Perspect Med. 2016, 6, a026179. [Google Scholar] [CrossRef]
- Laroche-Clary, A.; Chaire, V.; Algeo, M.P.; Derieppe, M.A.; Loarer, F.L.; Italiano, A. Combined targeting of MDM2 and CDK4 is synergistic in dedifferentiated liposarcomas. J. Hematol. Oncol. 2017, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.P.; Brown, C.J.; Verma, C.; Cheok, C.F. New insights into p53 based therapy. Discov. Med. 2011, 12, 107–117. [Google Scholar] [PubMed]
- Li, L.; Zhang, Y.; Gao, Y.; Hu, Y.; Wang, R.; Wang, S.; Li, Y.; He, Y.; Yuan, C. LncSNHG14 promotes nutlin3a resistance by inhibiting ferroptosis via the miR-206/SLC7A11 axis in osteosarcoma cells. Cancer Gene Ther. 2023, 30, 704–715. [Google Scholar] [CrossRef]
- Shen, J.K.; Cote, G.M.; Choy, E.; Yang, P.; Harmon, D.; Schwab, J.; Nielsen, G.P.; Chebib, I.; Ferrone, S.; Wang, X.; et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol. Res. 2014, 2, 690–698. [Google Scholar] [CrossRef]
- Cai, W.; Sun, Y.; Wang, W.; Han, C.; Ouchida, M.; Xia, W.; Zhao, X.; Sun, B. The effect of SYT-SSX and extracellular signal-regulated kinase (ERK) on cell proliferation in synovial sarcoma. Pathol. Oncol. Res. 2011, 17, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Kawai, A.; Araki, N.; Sugiura, H.; Ueda, T.; Yonemoto, T.; Takahashi, M.; Morioka, H.; Hiraga, H.; Hiruma, T.; Kunisada, T.; et al. Trabectedin monotherapy after standard chemotherapy versus best supportive care in patients with advanced, translocation-related sarcoma: A randomised, open-label, phase 2 study. Lancet Oncol. 2015, 16, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hao, M.; Du, X.; Chen, K.; Wang, G.; Yang, J. Advances in targeted therapy for osteosarcoma. Discov. Med. 2014, 17, 301–307. [Google Scholar]
- Anderson, J.L.; Park, A.; Akiyama, R.; Tap, W.D.; Denny, C.T.; Federman, N. Evaluation of In Vitro Activity of the Class I PI3K Inhibitor Buparlisib (BKM120) in Pediatric Bone and Soft Tissue Sarcomas. PLoS ONE 2015, 10, e0133610. [Google Scholar] [CrossRef]
- Babichev, Y.; Kabaroff, L.; Datti, A.; Uehling, D.; Isaac, M.; Al-Awar, R.; Prakesch, M.; Sun, R.X.; Boutros, P.C.; Venier, R.; et al. PI3K/AKT/mTOR inhibition in combination with doxorubicin is an effective therapy for leiomyosarcoma. J. Transl. Med. 2016, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Grignani, G.; Palmerini, E.; Ferraresi, V.; D’Ambrosio, L.; Bertulli, R.; Asaftei, S.D.; Tamburini, A.; Pignochino, Y.; Sangiolo, D.; Marchesi, E.; et al. Sorafenib and everolimus for patients with unresectable high-grade osteosarcoma progressing after standard treatment: A non-randomised phase 2 clinical trial. Lancet Oncol. 2015, 16, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.; Lee, J.; Rha, S.Y.; Park, K.H.; Kim, T.M.; Kim, Y.J.; Lee, H.J.; Lee, K.H.; Ahn, J.H. Multicenter phase II study of everolimus in patients with metastatic or recurrent bone and soft-tissue sarcomas after failure of anthracycline and ifosfamide. Invest. New Drugs 2013, 31, 1602–1608. [Google Scholar] [CrossRef]
- van der Graaf, W.T.; Blay, J.Y.; Chawla, S.P.; Kim, D.W.; Bui-Nguyen, B.; Casali, P.G.; Schoffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef]
- Ronellenfitsch, U.; Karampinis, I.; Dimitrakopoulou-Strauss, A.; Sachpekidis, C.; Jakob, J.; Kasper, B.; Nowak, K.; Pilz, L.; Attenberger, U.; Gaiser, T.; et al. Preoperative Pazopanib in High-Risk Soft Tissue Sarcoma: Phase II Window-of Opportunity Study of the German Interdisciplinary Sarcoma Group (NOPASS/GISG-04). Ann. Surg. Oncol. 2019, 26, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Gounder, M.M.; Mahoney, M.R.; Van Tine, B.A.; Ravi, V.; Attia, S.; Deshpande, H.A.; Gupta, A.A.; Milhem, M.M.; Conry, R.M.; Movva, S.; et al. Sorafenib for Advanced and Refractory Desmoid Tumors. N. Engl. J. Med. 2018, 379, 2417–2428. [Google Scholar] [CrossRef]
- D’Adamo, D.R.; Dickson, M.A.; Keohan, M.L.; Carvajal, R.D.; Hensley, M.L.; Hirst, C.M.; Ezeoke, M.O.; Ahn, L.; Qin, L.X.; Antonescu, C.R.; et al. A Phase II Trial of Sorafenib and Dacarbazine for Leiomyosarcoma, Synovial Sarcoma, and Malignant Peripheral Nerve Sheath Tumors. Oncologist 2019, 24, 857–863. [Google Scholar] [CrossRef]
- Maki, R.G.; D’Adamo, D.R.; Keohan, M.L.; Saulle, M.; Schuetze, S.M.; Undevia, S.D.; Livingston, M.B.; Cooney, M.M.; Hensley, M.L.; Mita, M.M.; et al. Phase II study of sorafenib in patients with metastatic or recurrent sarcomas. J. Clin. Oncol. 2009, 27, 3133–3140. [Google Scholar] [CrossRef]
- Que, Y.; Liang, Y.; Zhao, J.; Ding, Y.; Peng, R.; Guan, Y.; Zhang, X. Treatment-related adverse effects with pazopanib, sorafenib and sunitinib in patients with advanced soft tissue sarcoma: A pooled analysis. Cancer Manag. Res. 2018, 10, 2141–2150. [Google Scholar] [CrossRef]
- Deshpande, H.; Roman, S.; Thumar, J.; Sosa, J.A. Vandetanib (ZD6474) in the Treatment of Medullary Thyroid Cancer. Clin. Med. Insights Oncol. 2011, 5, 213–221. [Google Scholar] [CrossRef] [PubMed]
- van Oosterom, A.T.; Judson, I.; Verweij, J.; Stroobants, S.; Donato di Paola, E.; Dimitrijevic, S.; Martens, M.; Webb, A.; Sciot, R.; Van Glabbeke, M.; et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: A phase I study. Lancet 2001, 358, 1421–1423. [Google Scholar] [CrossRef]
- Kasper, B.; Gruenwald, V.; Reichardt, P.; Bauer, S.; Rauch, G.; Limprecht, R.; Sommer, M.; Dimitrakopoulou-Strauss, A.; Pilz, L.; Haller, F.; et al. Imatinib induces sustained progression arrest in RECIST progressive desmoid tumours: Final results of a phase II study of the German Interdisciplinary Sarcoma Group (GISG). Eur. J. Cancer 2017, 76, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Gounder, M.M.; Zer, A.; Tap, W.D.; Salah, S.; Dickson, M.A.; Gupta, A.A.; Keohan, M.L.; Loong, H.H.; D’Angelo, S.P.; Baker, S.; et al. Phase IB Study of Selinexor, a First-in-Class Inhibitor of Nuclear Export, in Patients With Advanced Refractory Bone or Soft Tissue Sarcoma. J. Clin. Oncol. 2016, 34, 3166–3174. [Google Scholar] [CrossRef]
- Duffaud, F.; Mir, O.; Boudou-Rouquette, P.; Piperno-Neumann, S.; Penel, N.; Bompas, E.; Delcambre, C.; Kalbacher, E.; Italiano, A.; Collard, O.; et al. Efficacy and safety of regorafenib in adult patients with metastatic osteosarcoma: A non-comparative, randomised, double-blind, placebo-controlled, phase 2 study. Lancet Oncol. 2019, 20, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Chuk, M.K.; Widemann, B.C.; Minard, C.G.; Liu, X.; Kim, A.; Bernhardt, M.B.; Kudgus, R.A.; Reid, J.M.; Voss, S.D.; Blaney, S.; et al. A phase 1 study of cabozantinib in children and adolescents with recurrent or refractory solid tumors, including CNS tumors: Trial ADVL1211, a report from the Children’s Oncology Group. Pediatr. Blood Cancer 2018, 65, e27077. [Google Scholar] [CrossRef] [PubMed]
- Gouda, M.A.; Nelson, B.E.; Buschhorn, L.; Wahida, A.; Subbiah, V. Tumor-Agnostic Precision Medicine from the AACR GENIE Database: Clinical implications. Clin. Cancer Res. 2023, 29, 2753–2760. [Google Scholar] [CrossRef]
- Hammer, K.J.; Copeland, V.C.; Loggers, E.T.; Pollack, S.M.; Wagner, M.J.; Cranmer, L.D. Doxorubicin and Olaratumab Versus Doxorubicin, Ifosfamide, and Mesna for Treatment of Advanced Soft Tissue Sarcomas. Am. J. Clin. Oncol. 2020, 43, 446–451. [Google Scholar] [CrossRef]
- Pappo, A.S.; Vassal, G.; Crowley, J.J.; Bolejack, V.; Hogendoorn, P.C.; Chugh, R.; Ladanyi, M.; Grippo, J.F.; Dall, G.; Staddon, A.P.; et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: Results of a Sarcoma Alliance. Cancer 2014, 120, 2448–2456. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Fujiwara, T.; Kunisada, T.; Ito, T.; Takihira, S.; Ozaki, T. Immunotherapy for sarcomas. Jpn J. Clin. Oncol. 2021, 51, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Sellars, M.C.; Wu, C.J.; Fritsch, E.F. Cancer vaccines: Building a bridge over troubled waters. Cell 2022, 185, 2770–2788. [Google Scholar] [CrossRef] [PubMed]
- Petitprez, F.; Reyniès, A.; Keung, E.Z.; Chen, T.; Sun, C.-M.; Calderaro, J.; Jeng, Y.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; de Angelis, A.; Ronchi, A.; De Chiara, A.; Fazioli, F.; Ruosi, C.; Altucci, L.; Conte, M.; de Nigris, F. Sarcoma Common MHC-I Haplotype Restricts Tumor-Specific CD8+ T Cell Response. Cancers 2022, 14, 3414. [Google Scholar] [CrossRef]
- Italiano, A.; Bellera, C.; D’Angelo, S. PD1/PD-L1 targeting in advanced soft-tissue sarcomas: A pooled analysis of phase II trials. J. Hematol. Oncol. 2020, 13, 55. [Google Scholar] [CrossRef]
- Molgora, M.; Esaulova, E.; Vermi, W.; Hou, J.; Chen, Y.; Luo, J.; Brioschi, S.; Bugatti, M.; Omodei, A.S.; Ricci, B.; et al. TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 2020, 182, 886–900.e817. [Google Scholar] [CrossRef]
- Maki, R.G.; Jungbluth, A.A.; Gnjatic, S.; Schwartz, G.K.; D’Adamo, D.R.; Keohan, M.L.; Wagner, M.J.; Scheu, K.; Chiu, R.; Ritter, E.; et al. A Pilot Study of Anti-CTLA4 Antibody Ipilimumab in Patients with Synovial Sarcoma. Sarcoma 2013, 2013, 168145. [Google Scholar] [CrossRef]
- Martin-Broto, J.; Hindi, N.; Grignani, G.; Martinez-Trufero, J.; Redondo, A.; Valverde, C.; Stacchiotti, S.; Lopez-Pousa, A.; D’Ambrosio, L.; Gutierrez, A.; et al. Nivolumab and sunitinib combination in advanced soft tissue sarcomas: A multicenter, single-arm, phase Ib/II trial. J. Immunother. Cancer 2020, 8, e001561. [Google Scholar] [CrossRef]
- Jackson, H.J.; Rafiq, S.; Brentjens, R.J. Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 2016, 13, 370–383. [Google Scholar] [CrossRef]
- Ahmed, N.; Mustafa, H.M.; Catrina, A.I.; Alstergren, P. Impact of temporomandibular joint pain in rheumatoid arthritis. Mediat. Inflamm. 2013, 2013, 597419. [Google Scholar] [CrossRef] [PubMed]
- D‘Angelo, S.P.; Melchiori, L.; Merchant, M.S.; Bernstein, D.; Glod, J.; Kaplan, R.; Grupp, S.; Tap, W.D.; Chagin, K.; Binder, G.K.; et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov. 2018, 8, 944–957. [Google Scholar] [CrossRef]
- Thanindratarn, P.; Dean, D.C.; Nelson, S.D.; Hornicek, F.J.; Duan, Z. Chimeric antigen receptor T (CAR-T) cell immunotherapy for sarcomas: From mechanisms to potential clinical applications. Cancer Treat. Rev. 2020, 82, 101934. [Google Scholar] [CrossRef] [PubMed]
- Concato, J.; Corrigan-Curay, J. Real-World Evidence—Where Are We Now? N. Engl. J. Med. 2022, 386, 1680–1682. [Google Scholar] [CrossRef]
- Videnovic, A.; Pfeiffer, H.C.V.; Tylki-Szymanska, A.; Berry-Kravis, E.; Ezgu, F.; Ganju, J.; Jurecka, A.; Lang, A.E. Study design challenges and strategies in clinical trials for rare diseases: Lessons learned from pantothenate kinase-associated neurodegeneration. Front. Neurol. 2023, 14, 1098454. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.; Demko, S.; Sinha, A.; Mishra-Kalyani, P.S.; Shen, Y.L.; Khasar, S.; Goheer, M.A.; Helms, W.S.; Pan, L.; Xu, Y.; et al. FDA Approval Summary: Selumetinib for Plexiform Neurofibroma. Clin. Cancer Res. 2021, 27, 4142–4146. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Ford, J.M.; O’Dwyer, P.J.; Shapiro, G.I.; McShane, L.M.; Freidlin, B.; O’Cearbhaill, R.E.; George, S.; Glade-Bender, J.; Lyman, G.H.; et al. National Cancer Institute Combination Therapy Platform Trial with Molecular Analysis for Therapy Choice (ComboMATCH). Clin. Cancer Res. 2023, 29, 1412–1422. [Google Scholar] [CrossRef]
- Lillie, E.O.; Patay, B.; Diamant, J.; Issell, B.; Topol, E.J.; Schork, N.J. The n-of-1 clinical trial: The ultimate strategy for individualizing medicine? Per. Med. 2011, 8, 161–173. [Google Scholar] [CrossRef]
- Subbiah, V.; Velcheti, V.; Tuch, B.B.; Ebata, K.; Busaidy, N.L.; Cabanillas, M.E.; Wirth, L.J.; Stock, S.; Smith, S.; Lauriault, V.; et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann. Oncol. 2018, 29, 1869–1876. [Google Scholar] [CrossRef]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef]
- Woodcock, J.; Marks, P. Drug Regulation in the Era of Individualized Therapies. N. Engl. J. Med. 2019, 381, 1678–1680. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Xi, R.; Wu, A.; Wang, S.; Li, Y.; Wang, C.; Tang, L.; Xia, Y.; Yang, D.; Li, J.; et al. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci. Transl. Med. 2020, 12, eaaz1723. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, G.; De Chiara, A.; Parafioriti, A.; Armiraglio, E.; Fazioli, F.; Gallo, M.; Aversa, L.; Camerlingo, R.; Cacciatore, F.; Colella, G.; et al. Patient-derived organoids as a potential model to predict response to PD-1/PD-L1 checkpoint inhibitors. Br. J. Cancer 2019, 121, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Pierrevelcin, M.; Flacher, V.; Mueller, C.G.; Vauchelles, R.; Guerin, E.; Lhermitte, B.; Pencreach, E.; Reisch, A.; Muller, Q.; Doumard, L.; et al. Engineering Novel 3D Models to Recreate High-Grade Osteosarcoma and its Immune and Extracellular Matrix Microenvironment. Adv. Heal. Mater. 2022, 11, e2200195. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.C.; Resasco, A.; Di Virgilio, A.L.; Ayala, M.; Cavaco, I.; Cabrera, S.; Aleman, J.; Leon, I.E. In vitro and in vivo anticancer effects of two quinoline-platinum(II) complexes on human osteosarcoma models. Cancer Chemother. Pharmacol. 2019, 83, 681–692. [Google Scholar] [CrossRef]
- Palubeckaite, I.; Venneker, S.; van den Akker, B.; Briaire-de Bruijn, I.H.; Bovee, J. Does PARP Inhibition Sensitize Chondrosarcoma Cell Lines to Chemotherapy or Radiotherapy? Results From a Three-dimensional Spheroid Cell Model. Clin. Orthop. Relat. Res. 2023, 481, 608–619. [Google Scholar] [CrossRef]
- Perrone, C.; Pomella, S.; Cassandri, M.; Pezzella, M.; Milano, G.M.; Colletti, M.; Cossetti, C.; Pericoli, G.; Di Giannatale, A.; de Billy, E.; et al. MET Inhibition Sensitizes Rhabdomyosarcoma Cells to NOTCH Signaling Suppression. Front. Oncol. 2022, 12, 835642. [Google Scholar] [CrossRef]
- Della Corte, C.M.; Barra, G.; Ciaramella, V.; Di Liello, R.; Vicidomini, G.; Zappavigna, S.; Luce, A.; Abate, M.; Fiorelli, A.; Caraglia, M.; et al. Antitumor activity of dual blockade of PD-L1 and MEK in NSCLC patients derived three-dimensional spheroid cultures. J. Exp. Clin. Cancer Res. 2019, 38, 253. [Google Scholar] [CrossRef]
- Wang, Y.; Pandey, R.N.; Roychoudhury, K.; Milewski, D.; Kalin, T.V.; Szabo, S.; Pressey, J.G.; Hegde, R.S. Targeting EYA3 in Ewing Sarcoma Retards Tumor Growth and Angiogenesis. Mol. Cancer Ther. 2021, 20, 803–815. [Google Scholar] [CrossRef]
- Fevre, R.; Mary, G.; Vertti-Quintero, N.; Durand, A.; Tomasi, R.F.; Del Nery, E.; Baroud, C.N. Combinatorial drug screening on 3D Ewing sarcoma spheroids using droplet-based microfluidics. iScience 2023, 26, 106651. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, R.; Yoshimatsu, Y.; Sin, Y.; Tsuchiya, R.; Ono, T.; Akiyama, T.; Hirabayashi, K.; Ozawa, I.; Nakagawa, R.; Kikuta, K.; et al. Establishment and characterization of two novel patient-derived myxoid liposarcoma cell lines. Hum. Cell 2022, 35, 1279–1289. [Google Scholar] [CrossRef]
- Aloy, M.T.; Sidi Boumedine, J.; Deville, A.; Kryza, D.; Gauthier, A.; Brichart-Vernos, D.; Ollier, G.; La Padula, V.; Lux, F.; Tillement, O.; et al. Proof of Concept of the Radiosensitizing Effect of Gadolinium Oxide Nanoparticles in Cell Spheroids and a Tumor-Implanted Murine Model of Chondrosarcoma. Int. J. Nanomed. 2022, 17, 6655–6673. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.; Truong, S.; Zhai, B.; Joshi, J.; Ghaidi, F.; Lizardo, M.M.; Shyp, T.; Kung, S.H.Y.; Rezakhanlou, A.M.; Oo, H.Z.; et al. A bi-functional PARP-HDAC inhibitor with activity in Ewing sarcoma. Clin. Cancer Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.R.; Lyle, A.G.; De Loose, A.; Maddukuri, L.; Learned, K.; Beale, H.C.; Kephart, E.T.; Cheney, A.; van den Bout, A.; Lee, M.P.; et al. A Functional Precision Medicine Pipeline Combines Comparative Transcriptomics and Tumor Organoid Modeling to Identify Bespoke Treatment Strategies for Glioblastoma. Cells 2021, 10, 3400. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Psilopatis, I.; Kokkali, S.; Palamaris, K.; Digklia, A.; Vrettou, K.; Theocharis, S. Organoids: A New Chapter in Sarcoma Diagnosis and Treatment. Int. J. Mol. Sci. 2022, 23, 11271. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef] [PubMed]
- Meister, M.T.; Groot Koerkamp, M.J.A.; de Souza, T.; Breunis, W.B.; Frazer-Mendelewska, E.; Brok, M.; DeMartino, J.; Manders, F.; Calandrini, C.; Kerstens, H.H.D.; et al. Mesenchymal tumor organoid models recapitulate rhabdomyosarcoma subtypes. EMBO Mol. Med. 2022, 14, e16001. [Google Scholar] [CrossRef]
- He, A.; Huang, Y.; Cheng, W.; Zhang, D.; He, W.; Bai, Y.; Gu, C.; Ma, Z.; He, Z.; Si, G.; et al. Organoid culture system for patient-derived lung metastatic osteosarcoma. Med. Oncol. 2020, 37, 105. [Google Scholar] [CrossRef]
- Al Shihabi, A.; Tebon, P.J.; Nguyen, H.T.L.; Chantharasamee, J.; Sartini, S.; Davarifar, A.; Jensen, A.Y.; Diaz-Infante, M.; Cox, H.; Enrique-Gonzalez, A.; et al. The landscape of drug sensitivity and resistance in sarcoma. bioRxiv 2023. [Google Scholar] [CrossRef]
- Forsythe, S.D.; Erali, R.A.; Sasikumar, S.; Laney, P.; Shelkey, E.; D’Agostino, R., Jr.; Miller, L.D.; Shen, P.; Levine, E.A.; Soker, S.; et al. Organoid Platform in Preclinical Investigation of Personalized Immunotherapy Efficacy in Appendiceal Cancer: Feasibility Study. Clin. Cancer Res. 2021, 27, 5141–5150. [Google Scholar] [CrossRef] [PubMed]
- Nagle, P.W.; Coppes, R.P. Current and Future Perspectives of the Use of Organoids in Radiobiology. Cells 2020, 9, 2649. [Google Scholar] [CrossRef]
- Roohani, S.; Loskutov, J.; Heufelder, J.; Ehret, F.; Wedeken, L.; Regenbrecht, M.; Sauer, R.; Zips, D.; Denker, A.; Joussen, A.M.; et al. Photon and Proton irradiation in Patient-derived, Three-Dimensional Soft Tissue Sarcoma Models. BMC Cancer 2023, 23, 577. [Google Scholar] [CrossRef]
- Forsythe, S.D.; Sivakumar, H.; Erali, R.A.; Wajih, N.; Li, W.; Shen, P.; Levine, E.A.; Miller, K.E.; Skardal, A.; Votanopoulos, K.I. Patient-Specific Sarcoma Organoids for Personalized Translational Research: Unification of the Operating Room with Rare Cancer Research and Clinical Implications. Ann. Surg. Oncol. 2022, 29, 7354–7367. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, X.; Ding, R.; Yang, L.; Lyu, X.; Zeng, J.; Lei, J.H.; Wang, L.; Bi, J.; Shao, N.; et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv. Sci. 2021, 8, e2101176. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, S.P.; Mahoney, M.R.; Van Tine, B.A.; Atkins, J.; Milhem, M.M.; Jahagirdar, B.N.; Antonescu, C.R.; Horvath, E.; Tap, W.D.; Schwartz, G.K.; et al. Nivolumab with or without ipilimumab treatment for metastatic sarcoma (Alliance A091401): Two open-label, non-comparative, randomised, phase 2 trials. Lancet Oncol. 2018, 19, 416–426. [Google Scholar] [CrossRef]
- Dajsakdipon, T.; Siripoon, T.; Ngamphaiboon, N.; Ativitavas, T.; Dejthevaporn, T. Immunotherapy and Biomarkers in Sarcoma. Curr. Treat. Options Oncol. 2022, 23, 415–438. [Google Scholar] [CrossRef]
- Xu, J.; Shi, Q.; Lou, J.; Wang, B.; Wang, W.; Niu, J.; Guo, L.; Chen, C.; Yu, Y.; Huang, Y.; et al. Chordoma recruits and polarizes tumor-associated macrophages via secreting CCL5 to promote malignant progression. J. Immunother. Cancer 2023, 11, e006808. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e1916. [Google Scholar] [CrossRef]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M.; et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer 2020, 122, 735–744. [Google Scholar] [CrossRef]
- Templeton, A.R.; Jeffery, P.L.; Thomas, P.B.; Perera, M.P.J.; Ng, G.; Calabrese, A.R.; Nicholls, C.; Mackenzie, N.J.; Wood, J.; Bray, L.J.; et al. Patient-Derived Explants as a Precision Medicine Patient-Proximal Testing Platform Informing Cancer Management. Front. Oncol. 2021, 11, 767697. [Google Scholar] [CrossRef]
- Nolan, J.; Pearce, O.M.T.; Screen, H.R.C.; Knight, M.M.; Verbruggen, S.W. Organ-on-a-Chip and Microfluidic Platforms for Oncology in the UK. Cancers 2023, 15, 635. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, M.; Robinson, S.; Alaei, B.; Clausen, M.; Hicks, R.; Belfield, G.; Althage, M.; Bak, A.; Lewis, J.A.; Hansen, P.B.L.; et al. 3D vascularised proximal tubules-on-a-multiplexed chip model for enhanced cell phenotypes. Lab Chip 2023, 23, 3226–3237. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, J.M.; Virumbrales-Munoz, M.; Lacueva, A.; Lanuza, P.M.; Checa-Chavarria, E.; Botella, P.; Fernandez, E.; Doblare, M.; Allison, S.J.; Phillips, R.M.; et al. Development and characterization of a microfluidic model of the tumour microenvironment. Sci. Rep. 2016, 6, 36086. [Google Scholar] [CrossRef] [PubMed]
- Majumder, B.; Baraneedharan, U.; Thiyagarajan, S.; Radhakrishnan, P.; Narasimhan, H.; Dhandapani, M.; Brijwani, N.; Pinto, D.D.; Prasath, A.; Shanthappa, B.U.; et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015, 6, 6169. [Google Scholar] [CrossRef]
- Pages, F.; Kirilovsky, A.; Mlecnik, B.; Asslaber, M.; Tosolini, M.; Bindea, G.; Lagorce, C.; Wind, P.; Marliot, F.; Bruneval, P.; et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 2009, 27, 5944–5951. [Google Scholar] [CrossRef]
- Voabil, P.; de Bruijn, M.; Roelofsen, L.M.; Hendriks, S.H.; Brokamp, S.; van den Braber, M.; Broeks, A.; Sanders, J.; Herzig, P.; Zippelius, A.; et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat. Med. 2021, 27, 1250–1261. [Google Scholar] [CrossRef]
- Kamer, I.; Bab-Dinitz, E.; Zadok, O.; Ofek, E.; Gottfried, T.; Daniel-Meshulam, I.; Hout-Siloni, G.; Ben Nun, A.; Barshack, I.; Onn, A.; et al. Immunotherapy response modeling by ex-vivo organ culture for lung cancer. Cancer Immunol. Immunother. 2021, 70, 2223–2234. [Google Scholar] [CrossRef]
- Ribas, A.; Shin, D.S.; Zaretsky, J.; Frederiksen, J.; Cornish, A.; Avramis, E.; Seja, E.; Kivork, C.; Siebert, J.; Kaplan-Lefko, P.; et al. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol. Res. 2016, 4, 194–203. [Google Scholar] [CrossRef]
- Mercatali, L.; Vanni, S.; Miserocchi, G.; Liverani, C.; Spadazzi, C.; Cocchi, C.; Calabrese, C.; Gurrieri, L.; Fausti, V.; Riva, N.; et al. The emerging role of cancer nanotechnology in the panorama of sarcoma. Front. Bioeng. Biotechnol. 2022, 10, 953555. [Google Scholar] [CrossRef]
- Chramiec, A.; Teles, D.; Yeager, K.; Marturano-Kruik, A.; Pak, J.; Chen, T.; Hao, L.; Wang, M.; Lock, R.; Tavakol, D.N.; et al. Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip 2020, 20, 4357–4372. [Google Scholar] [CrossRef] [PubMed]
- de Witte, C.J.; Espejo Valle-Inclan, J.; Hami, N.; Lohmussaar, K.; Kopper, O.; Vreuls, C.P.H.; Jonges, G.N.; van Diest, P.; Nguyen, L.; Clevers, H.; et al. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020, 31, 107762. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhao, H.; Zhang, W.; Wang, J.; Liu, Y.; Cao, Y.; Zheng, H.; Hu, Z.; Wang, S.; Zhu, Y.; et al. An Automated Organoid Platform with Inter-organoid Homogeneity and Inter-patient Heterogeneity. Cell Rep. Med. 2020, 1, 100161. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef]
- Vanni, S.; De Vita, A.; Gurrieri, L.; Fausti, V.; Miserocchi, G.; Spadazzi, C.; Liverani, C.; Cocchi, C.; Calabrese, C.; Bongiovanni, A.; et al. Myxofibrosarcoma landscape: Diagnostic pitfalls, clinical management and future perspectives. Ther. Adv. Med. Oncol. 2022, 14, 17588359221093973. [Google Scholar] [CrossRef]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef] [PubMed]



| Clinical Trials Including Organoid Platforms | |||||
|---|---|---|---|---|---|
| NCT Number | Study Title | Tumour | Intervention | Sample Size | Drug |
| NCT05890781 (recruiting) | Engineering Immune Organoids Study Pediatric Cancer | Sarcoma, brain tumour, kidney tumour, neuroblastoma | Fresh tumour sample to engineer immune organoids from paediatric patient tissues using induced pluripotent stem cells (iPSCs) | 100 | Not specified |
| NCT04986748 (recruiting) | Using QPOP to Predict Treatment for Sarcomas and Melanomas | Sarcomas and melanomas | Tumour samples for two- and three-dimensional models to evaluate drug sensitivities ex vivo | 100 | Panel of 14 drugs |
| NCT02910895 (recruiting) | A Platform of Patient Derived Xenografts (PDX) and 2D/3D Cell Cultures of Soft Tissue Sarcomas | Soft-tissue sarcoma | Sarcoma-patient-derived xenografts (sarcoma PDXs) | 54 | Not specified |
| NCT03358628 (not yet recruiting) | Patient-derived Xenograft (PDX) Modeling to Test Drug Response for High-grade Osteosarcoma | Osteosarcoma | Molecular profiling and in vivo drug testing in PDX | Unknown | |
| NCT03896958 (recruiting) | The PIONEER Initiative: Precision Insights On N-of-1 Ex Vivo Effectiveness Research Based on Individual Tumour Ownership (Precision Oncology) | Cancer | Functional precision testing of a patient’s tumour tissue to help guide optimal therapy (organoid, drug screening approaches in addition to traditional genomic profiling) | 1000 | Not specified |
| NCT05537844 (recruiting) | Longitudinal Sample Collection to Investigate Adaptation and Evolution of Ovarian High-grade Serous Carcinoma | Ovarian cancer (sarcoma, ovarian) | To acquire tumour material at diagnosis and relapse, whole blood for genomic analysis, plasma for ctDNA, and to isolate single cells and establish organoid cultures | 250 | Not specified |
| NCT04931381 (recruiting) | Organoid-Guided Chemotherapy for Advanced Pancreatic Cancer | Pancreatic cancer | Organoid test | 100 | Chemotherapy gemcitabine, 5-fluorouracil, paclitaxel, oxaliplatin, irinotecan |
| NCT04931394 (recruiting) | Organoid-Guided Adjuvant Chemotherapy for Pancreatic Cancer | Pancreatic cancer | Organoid drug test | 200 | Adjuvant chemotherapy |
| NCT05351398 (not yet recruiting) | The Clinical Efficacy of Drug Sensitive Neoadjuvant Chemotherapy Based on Organoid Versus Traditional Neoadjuvant Chemotherapy in Advanced Gastric Cancer | Advanced gastric cancer | Organoid drug test | 54 | Patient-derived, organoid-based, drug-sensitive, neoadjuvant chemotherapy |
| NCT05378048 (withdrawn) | Patient-derived-organoid (PDO) Guided Versus Conventional Therapy for Advanced Inoperable Abdominal Tumors | Advanced inoperable abdominal tumours | Organoid test | 140 | Not specified, genome-guided drug screening |
| NCT05352165 (not yet recruiting) | The Clinical Efficacy of Drug Sensitive Neoadjuvant Chemotherapy Based on Organoid Versus Traditional Neoadjuvant Chemotherapy in Advanced Rectal Cancer | Rectal cancer | Organoid drug test | 190 | Standard long-term therapy, wit the addition of FOLFOX; FOLFIRI; or 5-FU; or, 5-FU and pembrolizumab; and other individualised treatments |
| NCT05024734 (recruiting) | Guiding Instillation in Non Muscle-invasive Bladder Cancer Based on Drug Screens in Patient Derived Organoids | Bladder cancer | Organoid drug test | 30 | Epirubicin, mitomycin, gemcitabine, docetaxel |
| NCT05267912 (recruiting) | Prospective Multicenter Study Evaluating Feasibility and Efficacy of Tumor Organoid-based Precision Medicine in Patients With Advanced Refractory Cancers | Advanced, pretreated solid tumours | Organoid drug test | 1919 | Not specified, panel consisting of chemotherapy, hormonal therapy, targeted therapy |
| NCT04611035 (recruiting) | Q-GAIN (Using Qpop to Predict Treatment for GAstroIntestinal caNcer) | Gastrointestinal cancer | Organoid drug test | 100 | Panel of 14 drugs |
| NCT04450706 (recruiting) | Functional Precision Oncology for Metastatic Breast Cancer | Breast cancer HER2-negative, breast cancer | Organoid drug test | 15 | Not specified, individualised panels based on genomic analysis and NCCN guidelines |
| NCT04842006 (recruiting) | Systemic Neoadjuvant and Adjuvant Control by Precision Medicine in Rectal Cancer (SYNCOPE) | Colorectal cancer | Organoid drug test | 93 | Not specified neoadjuvant therapy and long radiation therapy |
| NCT05432518 (recruiting) | GBM Personalized Trial (Pilot Trial for Treatment of Recurrent Glioblastoma) | Recurrent glioblastoma | Organoid drug test | 10 | Afatinib, dasatinib, palbociclib, everolimus, olaparib |
| NCT05381038 (not yet recruiting) | Optimizing and Personalizing Azacitidine Combination Therapy for Treating Solid Tumors QPOP and CURATE.AI | Gastrointestinal cancer, breast Cancer | Organoids evaluated with QPOP | 10 | Azacitidine in combination with docetaxel, paclitaxel, or irinotecan |
| NCT05473923 (recruiting) | PTCs-based Precision Treatment Strategy on Recurrent High-grade Gliomas | Recurrent high-grade glioma | Patient-derived tumour-like cell clusters (multicellular spheroid model) | 30 | Non-specified, receiving chemotherapeutic or targeted drugs recommended by molecular tumour board |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Nigris, F.; Meo, C.; Palinski, W. Combination of Genomic Landsscape and 3D Culture Functional Assays Bridges Sarcoma Phenotype to Target and Immunotherapy. Cells 2023, 12, 2204. https://doi.org/10.3390/cells12172204
de Nigris F, Meo C, Palinski W. Combination of Genomic Landsscape and 3D Culture Functional Assays Bridges Sarcoma Phenotype to Target and Immunotherapy. Cells. 2023; 12(17):2204. https://doi.org/10.3390/cells12172204
Chicago/Turabian Stylede Nigris, Filomena, Concetta Meo, and Wulf Palinski. 2023. "Combination of Genomic Landsscape and 3D Culture Functional Assays Bridges Sarcoma Phenotype to Target and Immunotherapy" Cells 12, no. 17: 2204. https://doi.org/10.3390/cells12172204