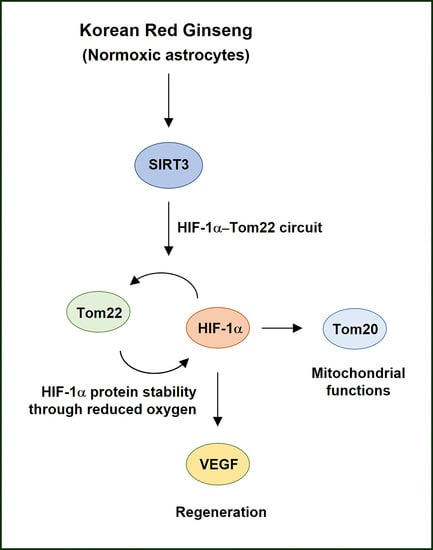
Korean Red Ginseng-Induced SIRT3 Promotes the Tom22–HIF-1α Circuit in Normoxic Astrocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals
2.3. Preparation of Brain Tissues for Immunohistochemistry
2.4. Primary Human Astrocyte Cell Culture
2.5. si-RNA Transfection
2.6. Immunocytochemistry
2.7. MitoTracker Staining
2.8. Western Blot Analysis
2.9. Oxidative Phosphorylation (OXPHOS)
2.10. Oxygen Consumption in Live Cells
2.11. Intracellular NAD+/NADH Measurement
2.12. Mitrochondria-Cytosol Fraction
2.13. ATP Levels in Astrocyte Cells
2.14. Protein Intensity in GFAP+-Astrocytes
2.15. Data Evaluation
3. Results
3.1. KRGE Induces VEGF in Astrocytes Located near the Corpus Callosum
3.2. KRGE Induces Astrocytic HIF-1α Protein Levels near the Corpus Callosum
3.3. KRGE-Induced SIRT3 Regulates HIF-1α Protein Levels
3.4. KRGE-Induced Nampt Did Not Upregulate NAD+/NADH Ratio or HIF-1α Proteins
3.5. HIF-1α-Mediated Expression of Mitochondria, Tom20, and Tom22 in KRGE-Treated Astrocytes
3.6. KRGE-Induced SIRT3 Regulates Astrocytic Mitochondrial Functions
3.7. KRGE Induces Astrocytic HIF-1α by Enhancing Tom22-Mediated Oxygen Consumption
3.8. KRGE-Mediated SIRT3 and Tom22 Stabilize HIF-1α Protein through PHD2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campello, L.; Singh, N.; Advani, J.; Mondal, A.K.; Corso-Diaz, X.; Swaroop, A. Aging of the retina: Molecular and metabolic turbulences and potential interventions. Annu. Rev. Vis. Sci. 2021, 7, 633–664. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Kim, J.H.; Lee, D.K.; Lee, K.S.; Won, M.H.; Jeoung, D.; Lee, H.; Ha, K.S.; Kwon, Y.G.; Kim, Y.M. Carbon monoxide potentiation of L-type Ca2+ channel activity increases HIF-1alpha-independent VEGF expression via an AMPKalpha/SIRT1-mediated PGC-1alpha/ERRalpha axis. Antioxid. Redox Signal. 2017, 27, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, M.; Kim, M.; Moon, S.; Kim, S.; Kim, S.; Koh, S.H.; Kim, Y.M.; Choi, Y.K. Prophylactic role of Korean Red Ginseng in astrocytic mitochondrial biogenesis through HIF-1alpha. J. Ginseng Res. 2022, 46, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Esposito, E.; Wang, X.; Terasaki, Y.; Liu, Y.; Xing, C.; Ji, X.; Lo, E.H. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016, 535, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, J.; Choi, Y.K. The role of astrocytes in the central nervous system focused on BK channel and heme oxygenase metabolites: A review. Antioxidants 2019, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.L.; Ousman, S.S. Astrocytes and Aging. Front. Aging Neurosci. 2018, 10, 337. [Google Scholar] [CrossRef]
- Araque, A. Astrocytes process synaptic information. Neuron Glia Biol. 2008, 4, 3–10. [Google Scholar] [CrossRef]
- Abbott, N.J.; Ronnback, L.; Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [Google Scholar] [CrossRef]
- Ryu, S.; Jeon, H.; Kim, H.Y.; Koo, S.; Kim, S. Korean red ginseng promotes hippocampal neurogenesis in mice. Neural Regen. Res. 2020, 15, 887–893. [Google Scholar] [CrossRef]
- Sung, W.N.; Kwok, H.H.; Rhee, M.H.; Yue, P.Y.; Wong, R.N. Korean Red Ginseng extract induces angiogenesis through activation of glucocorticoid receptor. J. Ginseng Res. 2017, 41, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.I.; Guarente, L. It takes two to tango: NAD(+) and sirtuins in aging/longevity control. npj Aging Mech. Dis. 2016, 2, 16017. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Tishkoff, D.X.; Bao, J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. In Histone Deacetylases: The Biology and Clinical Implication. Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 206, pp. 163–188. [Google Scholar] [CrossRef]
- Sidorova-Darmos, E.; Sommer, R.; Eubanks, J.H. The role of SIRT3 in the brain under physiological and pathological conditions. Front. Cell. Neurosci. 2018, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Sidorova-Darmos, E.; Wither, R.G.; Shulyakova, N.; Fisher, C.; Ratnam, M.; Aarts, M.; Lilge, L.; Monnier, P.P.; Eubanks, J.H. Differential expression of sirtuin family members in the developing, adult, and aged rat brain. Front. Aging Neurosci. 2014, 6, 333. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.R.; Semenza, G.L. Oxygen sensing and homeostasis. Physiology 2015, 30, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Park, J.H.; Yun, J.A.; Cha, J.H.; Kim, Y.; Won, M.H.; Kim, K.W.; Ha, K.S.; Kwon, Y.G.; Kim, Y.M. Heme oxygenase metabolites improve astrocytic mitochondrial function via a Ca2+-dependent HIF-1alpha/ERRalpha circuit. PLoS ONE 2018, 13, e0202039. [Google Scholar] [CrossRef]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Arany, Z.; Foo, S.Y.; Ma, Y.; Ruas, J.L.; Bommi-Reddy, A.; Girnun, G.; Cooper, M.; Laznik, D.; Chinsomboon, J.; Rangwala, S.M.; et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 2008, 451, 1008–1012. [Google Scholar] [CrossRef]
- Jin, K.; Zhu, Y.; Sun, Y.; Mao, X.O.; Xie, L.; Greenberg, D.A. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 11946–11950. [Google Scholar] [CrossRef]
- Gerhardt, H.; Golding, M.; Fruttiger, M.; Ruhrberg, C.; Lundkvist, A.; Abramsson, A.; Jeltsch, M.; Mitchell, C.; Alitalo, K.; Shima, D.; et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 2003, 161, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Pitt, A.S.; Buchanan, S.K. A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease. Cells 2021, 10, 1164. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.I.; Zhou, W. LKB1 regulated pathways in lung cancer invasion and metastasis. J. Thorac. Oncol. 2010, 5, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Moon, S.; Jeon, H.S.; Kim, S.; Koh, S.H.; Chang, M.S.; Kim, Y.M.; Choi, Y.K. Dual effects of Korean Red Ginseng on astrocytes and neural stem cells in traumatic brain injury: The HO-1-Tom20 axis as a putative target for mitochondrial function. Cells 2022, 11, 892. [Google Scholar] [CrossRef]
- Moon, S.; Chang, M.S.; Koh, S.H.; Choi, Y.K. Repair mechanisms of the neurovascular unit after ischemic stroke with a focus on VEGF. Int. J. Mol. Sci. 2021, 22, 8543. [Google Scholar] [CrossRef]
- Wright, G.L.; Maroulakou, I.G.; Eldridge, J.; Liby, T.L.; Sridharan, V.; Tsichlis, P.N.; Muise-Helmericks, R.C. VEGF stimulation of mitochondrial biogenesis: Requirement of AKT3 kinase. FASEB J. 2008, 22, 3264–3275. [Google Scholar] [CrossRef]
- Lin, J.B.; Kubota, S.; Ban, N.; Yoshida, M.; Santeford, A.; Sene, A.; Nakamura, R.; Zapata, N.; Kubota, M.; Tsubota, K.; et al. NAMPT-mediated NAD(+) biosynthesis is essential for vision in mice. Cell Rep. 2016, 17, 69–85. [Google Scholar] [CrossRef]
- Kobayashi, M.; Takeda, K.; Narita, T.; Nagai, K.; Okita, N.; Sudo, Y.; Miura, Y.; Tsumoto, H.; Nakagawa, Y.; Shimano, H.; et al. Mitochondrial intermediate peptidase is a novel regulator of sirtuin-3 activation by caloric restriction. FEBS Lett. 2017, 591, 4067–4073. [Google Scholar] [CrossRef]
- Appelhoff, R.J.; Tian, Y.M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef]
- Berra, E.; Benizri, E.; Ginouves, A.; Volmat, V.; Roux, D.; Pouyssegur, J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003, 22, 4082–4090. [Google Scholar] [CrossRef]
- Cheng, Y.; Shen, L.H.; Zhang, J.T. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action. Acta Pharmacol. Sin. 2005, 26, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, S.; Zhai, L.; Sun, L.; Zhao, D.; Wang, Z.; Li, X. Ginsenoside extract from ginseng extends lifespan and health span in Caenorhabditis elegans. Food Funct. 2021, 12, 6793–6808. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Kim, C.H.; Park, J.; Kim, M.; Jeon, H.S.; Kim, Y.M.; Choi, Y.K. Induction of BVR-A expression by Korean Red Ginseng in murine hippocampal astrocytes: Role of bilirubin in mitochondrial function via the LKB1-SIRT1-ERRalpha axis. Antioxidants 2022, 11, 1742. [Google Scholar] [CrossRef]
- Baranova, O.; Miranda, L.F.; Pichiule, P.; Dragatsis, I.; Johnson, R.S.; Chavez, J.C. Neuron-specific inactivation of the hypoxia inducible factor 1 alpha increases brain injury in a mouse model of transient focal cerebral ischemia. J. Neurosci. 2007, 27, 6320–6332. [Google Scholar] [CrossRef] [PubMed]
- Vecera, J.; Prochazkova, J.; Sumberova, V.; Panska, V.; Paculova, H.; Lanova, M.K.; Masek, J.; Bohaciakova, D.; Andersson, E.R.; Pachernik, J. Hypoxia/Hif1alpha prevents premature neuronal differentiation of neural stem cells through the activation of Hes1. Stem Cell Res. 2020, 45, 101770. [Google Scholar] [CrossRef]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD(+) metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.P.; He, Y.H.; Ding, J.C. Ginsenoside Rg1 Performs Anti-Aging Functions by Suppressing Mitochondrial Pathway-Mediated Apoptosis and Activating Sirtuin 3 (SIRT3)/Superoxide Dismutase 2 (SOD2) Pathway in Sca-1(+) HSC/HPC Cells of an Aging Rat Model. Med. Sci. Monit. 2020, 26, e920666. [Google Scholar] [CrossRef]
- Fan, C.; Ma, Q.; Xu, M.; Qiao, Y.; Zhang, Y.; Li, P.; Bi, Y.; Tang, M. Ginsenoside Rb1 Attenuates High Glucose-Induced Oxidative Injury via the NAD-PARP-SIRT Axis in Rat Retinal Capillary Endothelial Cells. Int. J. Mol. Sci. 2019, 20, 4936. [Google Scholar] [CrossRef]
- Sakaue, H.; Shiota, T.; Ishizaka, N.; Kawano, S.; Tamura, Y.; Tan, K.S.; Imai, K.; Motono, C.; Hirokawa, T.; Taki, K.; et al. Porin associates with Tom22 to regulate the mitochondrial protein gate assembly. Mol. Cell 2019, 73, 1044–1055.e8. [Google Scholar] [CrossRef]
- Mylonis, I.; Kourti, M.; Samiotaki, M.; Panayotou, G.; Simos, G. Mortalin-mediated and ERK-controlled targeting of HIF-1alpha to mitochondria confers resistance to apoptosis under hypoxia. J. Cell Sci. 2017, 130, 466–479. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Moon, S.; Lee, D.; Park, J.; Kim, C.-H.; Kim, Y.-M.; Choi, Y.K. Korean Red Ginseng-Induced SIRT3 Promotes the Tom22–HIF-1α Circuit in Normoxic Astrocytes. Cells 2023, 12, 1512. https://doi.org/10.3390/cells12111512
Kim H, Moon S, Lee D, Park J, Kim C-H, Kim Y-M, Choi YK. Korean Red Ginseng-Induced SIRT3 Promotes the Tom22–HIF-1α Circuit in Normoxic Astrocytes. Cells. 2023; 12(11):1512. https://doi.org/10.3390/cells12111512
Chicago/Turabian StyleKim, Hyungsu, Sunhong Moon, Dohyung Lee, Jinhong Park, Chang-Hee Kim, Young-Myeong Kim, and Yoon Kyung Choi. 2023. "Korean Red Ginseng-Induced SIRT3 Promotes the Tom22–HIF-1α Circuit in Normoxic Astrocytes" Cells 12, no. 11: 1512. https://doi.org/10.3390/cells12111512