The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle
Abstract
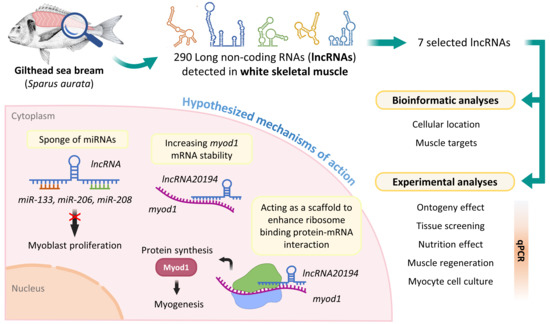
:1. Introduction
2. Materials and Methods
2.1. Identification of lncRNAs in Gilthead Sea Bream
2.2. RNA-Seq
2.3. Prediction of Target mRNAs, miRNAs, and Cell Location of lncRNAs
2.4. Experimental Trials
2.4.1. In Vivo Experiments
2.4.2. In Vitro Myogenesis: Primary Myocyte Cell Culture
2.5. Primer Design
2.6. Gene Expression
2.6.1. RNA Extraction and cDNA Synthesis
2.6.2. Quantitative Real-Time PCR (qPCR)
2.7. Statistics
3. Results
3.1. lncRNAs Selection
3.2. lncRNAs Subcellular Location and Targets
3.3. lncRNAs Expression in Gilthead Sea Bream Muscle
3.3.1. lncRNAs Expression in Fast Muscle of Fingerlings, Juveniles, and Adults of Gilthead Sea Bream
3.3.2. lncRNAs Tissue Screening
3.3.3. lncRNAs Expression in Response to Nutrition
3.3.4. lncRNAs Expression during Myogenesis
3.3.5. lncRNAs Expression during Muscle Regeneration
3.3.6. Correlation between lncRNAs and Genes Related to Muscle Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- APROMAR. APROMAR Report AQUACULTURE IN SPAIN. Available online: http://www.apromar.es/content/informes-anuales (accessed on 29 November 2021).
- Mommsen, T.P. Paradigms of growth in fish. Comp. Biochem. Physiol.—B Biochem. Mol. Biol. 2001, 129, 207–219. [Google Scholar] [CrossRef]
- Johnston, I.A. Environment and plasticity of myogenesis in teleost fish. J. Exp. Biol. 2006, 209, 2249–2264. [Google Scholar] [CrossRef] [PubMed]
- García de la serrana, D.; Codina, M.; Capilla, E.; Jiménez-Amilburu, V.; Navarro, I.; Du, S.J.; Johnston, I.A.; Gutiérrez, J. Characterisation and expression of myogenesis regulatory factors during in vitro myoblast development and in vivo fasting in the gilthead sea bream (Sparus aurata). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2014, 167, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A.; Garcia de la serrana, D.; Devlin, R.H. Muscle fibre size optimisation provides flexibility for energy budgeting in calorie-restricted coho salmon transgenic for growth hormone. J. Exp. Biol. 2014, 217, 3392–3395. [Google Scholar] [CrossRef]
- Lehka, L.; Rędowicz, M.J. Mechanisms regulating myoblast fusion: A multilevel interplay. Semin. Cell Dev. Biol. 2020, 104, 81–92. [Google Scholar] [CrossRef]
- Maître, J.L.; Heisenberg, C.P. Three Functions of Cadherins in Cell Adhesion. Curr. Biol. 2013, 23, R626. [Google Scholar] [CrossRef]
- Pfannkuche, K. Cell Fusion: Overviews and Methods, 2nd ed.; Springer: New York, NY, USA, 2015; pp. 1–248. [Google Scholar] [CrossRef]
- Leikina, E.; Gamage, D.G.; Prasad, V.; Goykhberg, J.; Crowe, M.; Diao, J.; Kozlov, M.M.; Chernomordik, L.V.; Millay, D.P. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell 2018, 46, 767–780.e7. [Google Scholar] [CrossRef]
- Perelló-Amorós, M.; Rallière, C.; Gutiérrez, J.; Gabillard, J.-C. Myomixer is expressed during embryonic and post-larval hyperplasia, muscle regeneration and differentiation of myoblasts in rainbow trout (Oncorhynchus mykiss). Gene 2021, 790, 145688. [Google Scholar] [CrossRef]
- Moore, C.A.; Parkin, C.A.; Bidet, Y.; Ingham, P.W. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 2007, 134, 3145–3153. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Tarifeño-Saldivia, E.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. In the shadow: The emerging role of long non-coding RNAs in the immune response of Atlantic salmon. Dev. Comp. Immunol. 2017, 73, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Non-coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, T.J.M.; Armand, A.S. Non-coding RNAs in skeletal muscle regeneration. Non-coding RNA Res. 2017, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Perelló-Amorós, M.; Otero-Tarrazón, A.; Jorge-Pedraza, V.; García-Pérez, I.; Sánchez-Moya, A.; Gabillard, J.C.; Moshayedi, F.; Navarro, I.; Capilla, E.; Fernández-Borràs, J.; et al. Myomaker and Myomixer characterization in gilthead sea bream under different myogenesis conditions. Sci. Rep. 2021; submitted. [Google Scholar]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Neguembor, M.V.; Jothi, M.; Gabellini, D. Long noncoding RNAs, emerging players in muscle\ndifferentiation and disease. Skelet. Muscle 2014, 4, 8. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA–LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Chen, S.; Li, W.; Chen, W.; Gu, W. LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding protein PTBP1. Oncogenesis 2019, 8, 73. [Google Scholar] [CrossRef]
- Chen, Y.; Wan, S.; Li, Q.; Dong, X.; Diao, J.; Liao, Q.; Wang, G.Y.; Gao, Z.X. Genome-Wide Integrated Analysis Revealed Functions of lncRNA–miRNA–mRNA Interaction in Growth of Intermuscular Bones in Megalobrama amblycephala. Front. Cell Dev. Biol. 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.N.; Wang, W.; Yang, N.; Huang, X.M.; Liu, C.F. Regulation of Glucose and Lipid Metabolism by Long Non-coding RNAs: Facts and Research Progress. Front. Endocrinol. 2020, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ding, C. Roles of LncRNAs in viral infections. Front. Cell. Infect. Microbiol. 2017, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Hadjicharalambous, M.R.; Lindsay, M.A. Long Non-Coding RNAs and the Innate Immune Response. Non-coding RNA 2019, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Deng, L.; Huang, N.; Sun, F. The Biological Roles of lncRNAs and Future Prospects in Clinical Application. Diseases 2021, 9, 8. [Google Scholar] [CrossRef]
- Vangoor, V.R.; Gomes-Duarte, A.; Pasterkamp, R.J. Long non-coding RNAs in motor neuron development and disease. J. Neurochem. 2021, 156, 777–801. [Google Scholar] [CrossRef]
- Sui, Y.; Han, Y.; Zhao, X.; Li, D.; Li, G. Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D. Cell Death Dis. 2019, 10, 181. [Google Scholar] [CrossRef]
- Wang, S.; Zuo, H.; Jin, J.; Lv, W.; Xu, Z.; Fan, Y.; Zhang, J.; Zuo, B. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 2019, 10, 505. [Google Scholar] [CrossRef]
- Wang, S.; Jin, J.; Xu, Z.; Zuo, B. Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells 2019, 8, 1107. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, M.; Lian, D.; Li, Y.; Li, Y.; Wang, J.; Deng, S.; Yu, K.; Lian, Z. Non-Coding RNA Regulates the Myogenesis of Skeletal Muscle Satellite Cells, Injury Repair and Diseases. Cells 2019, 8, 988. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed]
- Cichewicz, M.A.; Kiran, M.; Przanowska, R.K.; Sobierajska, E.; Shibata, Y.; Dutta, A. MUNC, an Enhancer RNA Upstream from the MYOD Gene, Induces a Subgroup of Myogenic Transcripts in trans Independently of MyoD. Mol. Cell. Biol. 2018, 38, e00655-17. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.C.; Cichewicz, M.A.; Dey, B.K.; Layer, R.; Reon, B.J.; Gagan, J.R.; Dutta, A. MUNC, a Long Noncoding RNA That Facilitates the Function of MyoD in Skeletal Myogenesis. Mol. Cell. Biol. 2015, 35, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Al-Tobasei, R.; Kenney, B.; Leeds, T.D.; Salem, M. Integrated analysis of lncRNA and mRNA expression in rainbow trout families showing variation in muscle growth and fillet quality traits. Sci. Rep. 2018, 8, 12111. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, L.; Yin, H.; Zhu, W.; Fu, J.; Dong, Z. Integrated analysis of long non-coding RNA and mRNA expression in different colored skin of koi carp. BMC Genomics 2019, 20, 515. [Google Scholar] [CrossRef]
- Wei, S.; Chen, Y.; Huang, L.; Ma, H.; Qi, L.; Wang, Q.; Sun, M.; Zhang, X.; Sha, Z. Analysis of lncRNA and mRNA expression profiles in peripheral blood leukocytes of the half-smooth tongue sole (Cynoglossus semilaevis) treated with chitosan oligosaccharide. Dev. Comp. Immunol. 2021, 120, 104043. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, W.; Zhu, A.; Zhang, J.; Zhou, J.; Wu, C. Transcriptome analysis demonstrate widespread differential expression of long noncoding RNAs involve in Larimichthys crocea immune response. Fish Shellfish Immunol. 2016, 51, 1–8. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wu, W.; Yu, F.; Chang, W.; Li, P.; Wang, K. Non-coding RNAs function as immune regulators in teleost fish. Front. Immunol. 2018, 9, 2801. [Google Scholar] [CrossRef]
- Cai, J.; Li, L.; Song, L.; Xie, L.; Luo, F.; Sun, S.; Chakraborty, T.; Zhou, L.; Wang, D. Effects of long term antiprogestine mifepristone (RU486) exposure on sexually dimorphic lncRNA expression and gonadal masculinization in Nile tilapia (Oreochromis niloticus). Aquat. Toxicol. 2019, 215, 105289. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, S.; Sun, D.; Wu, Z.; Wei, C.; Dai, C.; Jiang, L.; Peng, S. Transcriptome profiling analysis of sex-based differentially expressed mRNAs and lncRNAs in the brains of mature zebrafish (Danio rerio). BMC Genomics 2019, 20, 830. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Váldes, J.A.; Gallardo-Escárate, C. Transcriptome Profiling of Long Non-coding RNAs During the Atlantic Salmon Smoltification Process. Mar. Biotechnol. 2021, 23, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Acuña, G.; Détrée, C.; Gallardo-Escárate, C.; Gonçalves, A.T. Functional Diets Modulate lncRNA-Coding RNAs and Gene Interactions in the Intestine of Rainbow Trout Oncorhynchus mykiss. Mar. Biotechnol. 2017, 19, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cao, L.; Sun, B.; Wei, Y.; Liang, M. Transcriptomic analysis of potential “lncRNA-mRNA” interactions in liver of the marine teleost Cynoglossus semilaevis fed diets with different DHA/EPA ratios. Front. Physiol. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Paneru, B.; Ali, A.; Al-Tobasei, R.; Kenney, B.; Salem, M. Crosstalk among lncRNAs, microRNAs and mRNAs in the muscle ‘degradome’ of rainbow trout. Sci. Rep. 2018, 8, 8416. [Google Scholar] [CrossRef] [PubMed]
- Lavajoo, F.; Perelló-Amorós, M.; Vélez, E.J.; Sánchez-Moya, A.; Balbuena-Pecino, S.; Riera-Heredia, N.; Fernández-Borràs, J.; Blasco, J.; Navarro, I.; Capilla, E.; et al. Regulatory mechanisms involved in muscle and bone remodeling during refeeding in gilthead sea bream. Sci. Rep. 2020, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Perelló-Amorós, M.; García-Pérez, I.; Sánchez-Moya, A.; Innamorati, A.; Vélez, E.J.; Achaerandio, I.; Pujolà, M.; Calduch-Giner, J.; Pérez-Sánchez, J.; Fernández-Borràs, J.; et al. Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata). Animals 2021, 11, 2182. [Google Scholar] [CrossRef]
- Sánchez-Moya, A.; García-Meilán, I.; Riera-Heredia, N.; Vélez, E.J.; Lutfi, E.; Fontanillas, R.; Gutiérrez, J.; Capilla, E.; Navarro, I. Effects of different dietary vegetable oils on growth and intestinal performance, lipid metabolism and flesh quality in gilthead sea bream. Aquaculture 2020, 519, 734881. [Google Scholar] [CrossRef]
- Vélez, E.J.; Azizi, S.; Millán-Cubillo, A.; Fernández-Borràs, J.; Blasco, J.; Chan, S.J.; Calduch-Giner, J.A.; Pérez-Sánchez, J.; Navarro, I.; Capilla, E.; et al. Effects of sustained exercise on GH-IGFs axis in gilthead sea bream (Sparus aurata). Am. J. Physiol.—Regul. Integr. Comp. Physiol. 2016, 310, R313–R322. [Google Scholar] [CrossRef]
- Vélez, E.J.; Azizi, S.; Verheyden, D.; Salmerón, C.; Lutfi, E.; Sánchez-Moya, A.; Navarro, I.; Gutiérrez, J.; Capilla, E. Proteolytic systems’ expression during myogenesis and transcriptional regulation by amino acids in gilthead sea bream cultured muscle cells. PLoS ONE 2017, 12, e0187339. [Google Scholar] [CrossRef]
- Naya-Català, F.; Martos-Sitcha, J.A.; de las Heras, V.; Simó-Mirabet, P.; Calduch-Giner, J.; Pérez-Sánchez, J. Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles. Biology 2021, 10, 416. [Google Scholar] [CrossRef]
- Balbuena-Pecino, S.; Riera-Heredia, N.; Vélez, E.J.; Gutiérrez, J.; Navarro, I.; Riera-Codina, M.; Capilla, E. Temperature Affects Musculoskeletal Development and Muscle Lipid Metabolism of Gilthead Sea Bream (Sparus aurata). Front. Endocrinol. 2019, 10, 173. [Google Scholar] [CrossRef] [PubMed]
- Garcia de la serrana, D.; Estévez, A.; Andree, K.; Johnston, I. a Fast skeletal muscle transcriptome of the gilthead sea bream (Sparus aurata) determined by next generation sequencing. BMC Genomics 2012, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Guidelines for the treatment of animals in behavioural research and teaching. Anim. Behav. 2000, 59, 253–257. [Google Scholar] [CrossRef]
- Metcalfe, J.D.; Craig, J.F. Ethical justification for the use and treatment of fishes in research: An update. J. Fish Biol. 2011, 78, 393–394. [Google Scholar] [CrossRef]
- Zhao, Q.-Y.; Wang, Y.; Kong, Y.-M.; Luo, D.; Li, X.; Hao, P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: A comparative study. BMC Bioinformatics 2011, 12, S2. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806–812. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Cao, Z.; Pan, X.; Yang, Y.; Huang, Y.; Shen, H.-B. The lncLocator: A subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 2018, 34, 2185–2194. [Google Scholar] [CrossRef]
- Perelló-Amorós, M.; Vélez, E.J.; Vela-Albesa, J.; Sánchez-Moya, A.; Riera-Heredia, N.; Hedén, I.; Fernández-Borràs, J.; Blasco, J.; Calduch-Giner, J.A.; Navarro, I.; et al. Ghrelin and its receptors in gilthead sea bream: Nutritional regulation. Front. Endocrinol. 2018, 9, 399. [Google Scholar] [CrossRef] [PubMed]
- Montserrat, N.; Sánchez-Gurmaches, J.; García De La serrana, D.; Navarro, I.; Gutiérrez, J. IGF-I binding and receptor signal transduction in primary cell culture of muscle cells of gilthead sea bream: Changes throughout in vitro development. Cell Tissue Res. 2007, 330, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 29 November 2021).
- Wickham, H. Ggplot2: Elegrant graphics for data analysis, 2nd ed.; Springer: New York, NY, USA, 2016; ISBN 9783319242750. [Google Scholar]
- Otero-Tarrazón, A.; Perelló-Amorós, M.; Sánchez-Moya, A.; Jorge-Pedraza, V.; Moshayedi, F.; García-Pérez, I.; Capilla, E.; Navarro, I.; Fernández-Borràs, J.; Garcia de la serrana, D.; et al. Muscle regeneration in gilthead sea bream: Implications of autocrine GH-IGF axis, the muscle proteolytic systems, novel regulatory factors, and the crosstalk with bone. Front. Endocrinol. 2022; in preparation. [Google Scholar]
- Chillón, I.; Marcia, M. The molecular structure of long non-coding RNAs: Emerging patterns and functional implications. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 662–690. [Google Scholar] [CrossRef]
- Zimmer-Bensch, G. Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8, 1399. [Google Scholar] [CrossRef]
- Altringham, J.D.; Ellerby, D.J. Fish swimming: Patterns in muscle function. J. Exp. Biol. 1999, 202, 3397–3403. [Google Scholar] [CrossRef]
- Amaral, I.P.G.; Johnston, I.A. Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. AJP Regul. Integr. Comp. Physiol. 2012, 302, R193–R206. [Google Scholar] [CrossRef]
- Garcia de la serrana, D.; Vieira, V.L.A.; Andree, K.B.; Darias, M.; Estévez, A.; Gisbert, E.; Johnston, I.A. Development Temperature Has Persistent Effects on Muscle Growth Responses in Gilthead Sea Bream. PLoS ONE 2012, 7, e51884. [Google Scholar] [CrossRef]
- Hagen, Ø.; Fernandes, J.M.O.; Solberg, C.; Johnston, I.A. Expression of growth-related genes in muscle during fasting and refeeding of juvenile Atlantic halibut, Hippoglossus hippoglossus L. Comp. Biochem. Physiol.—B Biochem. Mol. Biol. 2009, 152, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.L.; Mann, C.J.; Vidal, B.; Ardite, E.; Perdiguero, E.; Muñoz-Cánoves, P. Cellular and Molecular Mechanisms Regulating Fibrosis in Skeletal Muscle Repair and Disease. Curr. Top. Dev. Biol. 2011, 96, 167–201. [Google Scholar] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite Cells and the Muscle Stem Cell Niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed]
- Ballarino, M.; Cazzella, V.; D’Andrea, D.; Grassi, L.; Bisceglie, L.; Cipriano, A.; Santini, T.; Pinnarò, C.; Morlando, M.; Tramontano, A.; et al. Novel long noncoding RNAs (lncRNAs) in myogenesis: A miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 2015, 35, 728–736. [Google Scholar] [CrossRef]
- Hitachi, K.; Nakatani, M.; Takasaki, A.; Ouchi, Y.; Uezumi, A.; Ageta, H.; Inagaki, H.; Kurahashi, H.; Tsuchida, K. Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 2019, 20, e47468. [Google Scholar] [CrossRef]
- Gong, C.; Li, Z.; Ramanujan, K.; Clay, I.; Zhang, Y.; Lemire-Brachat, S.; Glass, D.J. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell 2015, 34, 181–191. [Google Scholar] [CrossRef]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Kang, M.-J.; Guo, R.; Kim, J.; Grammatikakis, I.; Yoon, J.-H.; Dudekula, D.B.; Noh, J.H.; Yang, X.; et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014, 42, 10099–10111. [Google Scholar] [CrossRef]
- Xu, H.; Jiang, Y.; Xu, X.; Su, X.; Liu, Y.; Ma, Y.; Zhao, Y.; Shen, Z.; Huang, B.; Cao, X. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019, 20, 1621–1630. [Google Scholar] [CrossRef]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the Cytoplasm. Genomics. Proteomics Bioinformatics 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Winbanks, C.E.; Ooi, J.Y.; Nguyen, S.S.; Mcmullen, J.R.; Bernardo, B.C. MicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: Potential drug targets? Clin. Exp. Pharmacol. Physiol. 2014, 41, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Mok, G.F.; Lozano-Velasco, E.; Münsterberg, A. microRNAs in skeletal muscle development. Semin. Cell Dev. Biol. 2017, 72, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Zhao, J.; Hu, G. Genome-wide methods for investigating long noncoding RNAs. Biomed. Pharmacother. 2019, 111, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Schönberger, B.; Schaal, C.; Schäfer, R.; Voß, B. RNA interactomics: Recent advances and remaining challenges. F1000Research 2018, 7, F1000. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Jensen, E. Technical review: In situ hybridization. Anat. Rec. 2014, 297, 1349–1353. [Google Scholar] [CrossRef]








| LncRNAs | mRNA | ndG | miRNA | MEF (kcal) | Subcellular Location | Probability |
|---|---|---|---|---|---|---|
| lncRNA14696 | lamtor2 (ENSSAUG00010013916) | −0.087 | miR-133a1/2 | −25.9 | Nucleus | 0.93 |
| miR-133b | −29.2 | |||||
| miR-499 | −20.6 | |||||
| miR-1 | −21.9 | |||||
| miR-206 | −28.3 | |||||
| miR-208 | −22.3 | |||||
| lncRNA16861 | lamtor1 (ENSSAUG00010010852) | −0.087 | miR-133b | −23.2 | Cytoplasm | 0.89 |
| miR-488 | −21.3 | |||||
| miR-1 | −21.4 | |||||
| miR-206 | −23.3 | |||||
| miR-208 | −21.3 | |||||
| lncRNA20194 | myod1 (ENSSAUG00010008630) | −0.170 | miR-133a1/2 | −23.4 | Cytoplasm | 0.82 |
| miR-133b | −26.2 | |||||
| miR-499 | −22.5 | |||||
| miR-206 | −21.1 | |||||
| miR-208 | −28.3 | |||||
| lncRNA21817 | NA | NA | miR-133a1/2 | −23.3 | Cytoplasm | 0.70 |
| miR-133b | −27.3 | |||||
| miR-499 | −23.4 | |||||
| miR-206 | −27.2 | |||||
| miR-208 | −21.6 | |||||
| lncRNA40141 | eif4ebp1 (ENSSAUG00010019173) | −0.085 | miR-133a1/2 | −22.4 | Cytoplasm | 0.46 |
| miR-133b | −25.8 | Nucleus | 0.26 | |||
| miR-499 | −20.5 | |||||
| miR-1 | −22.0 | |||||
| miR-206 | −25.2 | |||||
| miR-208 | −26.1 | |||||
| lncRNA54283 | lamtor1 (ENSSAUG00010010852) | −0.082 | miR-133b | −23.4 | Nucleus | 0.49 |
| pax7b (ENSSAUG00010013590) | −0.080 | miR-206 | −24.3 | Cytoplasm | 0.37 | |
| miR-208 | −21.9 | |||||
| lncRNA60660 | NA | NA | miR-206 | −25.1 | Nucleus | 0.67 |
| Cytoplasm | 0.27 |
| lncRNAs | myf5 | myod1 | myod2 | myog | mef2c | myf6 | cdh15 | cav3 | mymx | mymk | dock5 | crk-a | crk-b | crkl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lncRNA14696 | 0.2 | 0.1 | −0.1 | −0.2 | 0.4 * | −0.1 | −0.1 | 0.3 | −0.1 | 0.1 | 0.5 ** | 0.3 | 0.1 | 0.4 * |
| lncRNA16861 | 0 | −0.1 | −0.1 | 0.1 | −0.1 | 0.1 | 0.1 | −0.4 * | 0.1 | −0.4 *** | 0.1 | −0.02 | 0.3 | −0.1 |
| lncRNA20194 | 0.0 | 0.6 *** | 0.1 | −0.4 * | 0.2 | 0.4 *** | −0.2 | 0.0 | 0.6 *** | 0.3 * | 0.4 ** | 0.4 * | 0.4 * | 0.2 |
| lncRNA21817 | 0 | 0.1 | −0.4 * | −0.5 ** | 0 | 0.1 | −0.4 * | 0.1 | −0.4 * | 0.1 | 0.6 ** | 0.4 * | 0.4 * | 0.3 |
| lncRNA40141 | −0.3 | 0.5 ** | −0.5 ** | −0.2 | −0.2 | −0.2 | −0.1 | −0.1 | −0.2 | −0.3 | 0.7 *** | 0.6 *** | 0.8 *** | 0.4 * |
| lncRNA54283 | −0.1 | 0.3 * | 0 | −0.1 | 0 | 0.1 | 0 | −0.6 ** | 0.3 * | −0.1 | 0.5 *** | 0.5** | 0.6 *** | 0.4 * |
| lncRNA60660 | 0.2 | 0.2 | −0.2 | −0.4 * | 0.0 | −0.4 * | −0.2 | −0.1 | −0.3 | 0.1 | 0.7 *** | 0.7 *** | 0.4 ** | 0.6 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Pérez, I.; Molsosa-Solanas, A.; Perelló-Amorós, M.; Sarropoulou, E.; Blasco, J.; Gutiérrez, J.; Garcia de la serrana, D. The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle. Cells 2022, 11, 428. https://doi.org/10.3390/cells11030428
García-Pérez I, Molsosa-Solanas A, Perelló-Amorós M, Sarropoulou E, Blasco J, Gutiérrez J, Garcia de la serrana D. The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle. Cells. 2022; 11(3):428. https://doi.org/10.3390/cells11030428
Chicago/Turabian StyleGarcía-Pérez, Isabel, Anna Molsosa-Solanas, Miquel Perelló-Amorós, Elena Sarropoulou, Josefina Blasco, Joaquim Gutiérrez, and Daniel Garcia de la serrana. 2022. "The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle" Cells 11, no. 3: 428. https://doi.org/10.3390/cells11030428