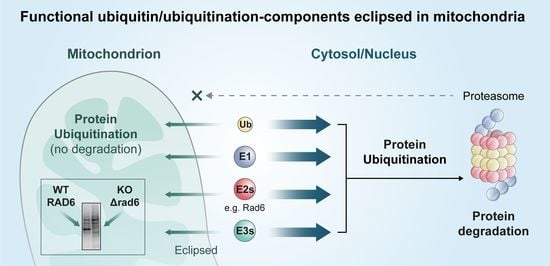
Ubiquitination Occurs in the Mitochondrial Matrix by Eclipsed Targeted Components of the Ubiquitination Machinery
Abstract
:1. Introduction
2. Methods
2.1. Growth Conditions of Yeast Cells
2.2. α-Complementation Assay
2.3. Subcellular Fractionation
2.4. Yeast Oxygen Consumption Rate Measurements Using Seahorse XFe96 Analyzer
2.5. Immunoprecipitation
2.6. Mitochondrial Trypsinization
2.7. MG132 Induced Proteasome Inhibition of Living Yeast Cells
2.8. Mass Spectrometry (MS)
3. Results
3.1. The α-Complementation Assay Indicates Targeting of Components of Ubiquitination System to Mitochondria of S. cerevisiae
3.2. Targeting HA-Tagged Ubiquitin to S. cerevisiae Mitochondria
3.2.1. Conjugates of MTS-Directed HA-Ub Are Detected Only in Mitochondrial Fractions
3.2.2. Conjugate Signals of preSu9-HA-Ub in Isolated Mitochondria Are Resistant to External Trypsin Treatment
3.2.3. preSu9-HA-Ub Conjugates Levels in Mitochondria Are Not Elevated by Proteasome Inhibition
3.2.4. Immunoprecipitation (IP) of preSu9-HA-Ub Conjugates Is Enriched in Mitochondrial Matrix Proteins
3.3. RAD6 Deficiency Causes an Altered Ubiquitination Pattern in Mitochondria
3.4. Rad6 Is Active in Mitochondria
3.5. Rad6 UBC Activity Is Required for Protein Ubiquitination in Mitochondria
3.6. The Rad6 N Terminus Is Required for Its Targeting to and Activity in Mitochondria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neutzner, M.; Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012, 52, 37–50. [Google Scholar] [PubMed] [Green Version]
- Chen, Z.J.; Sun, L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [Green Version]
- Picard, M.; Wallace, D.C.; Burelle, Y. The rise of mitochondria in medicine. Mitochondrion 2016, 30, 105–116. [Google Scholar] [CrossRef]
- Malina, C.; Larsson, C.; Nielsen, J. Yeast mitochondria: An overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 2018, 18, foy040. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livnat-Levanon, N.; Glickman, M.H. Ubiquitin-proteasome system and mitochondria—Reciprocity. Biochim. Biophys. Acta 2011, 1809, 80–87. [Google Scholar] [CrossRef]
- Sulkshane, P.; Ram, J.; Glickman, M.H. Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability. Biomolecules 2020, 10, 1559. [Google Scholar] [CrossRef]
- von Mikecz, A. The nuclear ubiquitin-proteasome system. J. Cell Sci. 2006, 119, 1977–1984. [Google Scholar] [CrossRef] [Green Version]
- Karniely, S.; Rayzner, A.; Sass, E.; Pines, O. Alpha-complementation as a probe for dual localization of mitochondrial proteins. Exp. Cell Res. 2006, 312, 3835–3846. [Google Scholar] [CrossRef]
- Ben-Menachem, R.; Pines, O. Detection of Dual Targeting and Dual Function of Mitochondrial Proteins in Yeast. Methods Mol Biol. 2017, 1567, 179–195. [Google Scholar]
- Regev-Rudzki, N.; Karniely, S.; Ben-Haim, N.N.; Pines, O. Yeast aconitase in two locations and two metabolic pathways: Seeing small amounts is believing. Mol. Biol. Cell 2005, 16, 4163–4171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regev-Rudzki, N.; Pines, O. Eclipsed distribution: A phenomenon of dual targeting of protein and its significance. Bioessays 2007, 29, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, G.; Udasin, R.G.; Ciechanover, A. On the linkage between the ubiquitin-proteasome system and the mitochondria. Biochem. Biophys. Res. Commun. 2016, 473, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, G.; Ziv, T.; Braten, O.; Admon, A.; Udasin, R.G.; Ciechanover, A. Ubiquitination of specific mitochondrial matrix proteins. Biochem. Biophys. Res. Commun. 2016, 475, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Lavie, J.; De Belvalet, H.; Sonon, S.; Ion, A.M.; Dumon, E.; Melser, S.; Lacombe, D.; Dupuy, J.W.; Lalou, C.; Bénard, G. Ubiquitin-Dependent Degradation of Mitochondrial Proteins Regulates Energy Metabolism. Cell Rep. 2018, 23, 2852–2863. [Google Scholar] [CrossRef] [PubMed]
- Daum, G.; Bohni, P.C.; Schatz, G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982, 257, 13028–13033. [Google Scholar] [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Schmidt, B.; Wachter, E.; Sebald, W.; Neupert, W. Processing peptidase of Neurospora mitochondria. Two-step cleavage of imported ATPase subunit 9. Eur. J. Biochem. 1984, 144, 581–588. [Google Scholar] [CrossRef] [Green Version]
- Hartl, F.U.; Pfanner, N.; Nicholson, D.W.; Neupert, W. Mitochondrial protein import. Biochim. Biophys. Acta 1989, 988, 1–45. [Google Scholar] [CrossRef] [Green Version]
- Westermann, B.; Neupert, W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 2000, 16, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Gomez, T.A.; Deshaies, R.J.; Tansey, W.P. Combined chemical and genetic approach to inhibit proteolysis by the proteasome. Yeast 2010, 27, 965–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vögtle, F.; Burkhart, J.M.; Gonczarowska-Jorge, H.; Kücükköse, C.; Taskin, A.A.; Kopczynski, D.; Ahrends, R.; Mossmann, D.; Sickmann, A.; Zahedi, R.P.; et al. Landscape of submitochondrial protein distribution. Nat. Commun. 2017, 8, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [Green Version]
- Bailly, V.; Lamb, J.; Sung, P.; Prakash, S.; Prakash, L. Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994, 8, 811–820. [Google Scholar] [CrossRef] [Green Version]
- Game, J.C.; Chernikova, S.B. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 2009, 8, 470–482. [Google Scholar] [CrossRef]
- Sung, P.; Berleth, E.; Pickart, C.; Prakash, S.; Prakash, L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 1991, 10, 2187–2193. [Google Scholar] [CrossRef]
- Stolz, A.; Besser, S.; Hottmann, H.; Wolf, D.H. Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 15271–15276. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Mao, X.; Ju, D.; Xie, Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 2004, 279, 55218–55223. [Google Scholar] [CrossRef] [Green Version]
- Ju, D.; Wang, X.; Xu, H.; Xie, Y. Genome-wide analysis identifies MYND-domain protein Mub1 as an essential factor for Rpn4 ubiquitylation. Mol. Cell Biol. 2008, 28, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Gilon, T.; Chomsky, O.; Kulka, R.G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998, 17, 2759–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robzyk, K.; Recht, J.; Osley, M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000, 287, 501–504. [Google Scholar] [CrossRef]
- Sung, P.; Prakash, S.; Prakash, L. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. USA 1990, 87, 2695–2699. [Google Scholar] [CrossRef] [Green Version]
- Broomfield, S.; Chow, B.L.; Xiao, W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5678–5683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, J.F.; Sung, P.; Prakash, S.; Prakash, L. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev. 1993, 7, 250–261. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.M.; Sedman, J.; Neupert, W.; Stuart, R.A. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem. 1999, 274, 20937–20942. [Google Scholar] [CrossRef]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [Green Version]
- Grau-Bove, X.; Sebe-Pedros, A.; Ruiz-Trillo, I. The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin. Mol. Biol. Evol. 2015, 32, 726–739. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Herrmann, J.M.; Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2021, 22, 54–70. [Google Scholar] [CrossRef]
- Kim, J.; Roeder, R.G. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 2009, 284, 20582–20592. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Magala, P.; Geiger-Schuller, K.R.; Majumdar, A.; Tolman, J.R.; Wolberger, C. Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity. Nucleic Acids Res. 2015, 43, 9039–9050. [Google Scholar] [CrossRef] [PubMed]



 ), an outer membrane protein exposed to trypsin (
), an outer membrane protein exposed to trypsin ( ), a matrix protein protected inside mitochondria (
), a matrix protein protected inside mitochondria ( ), and trypsin (
), and trypsin ( ) are indicated. (D) preSu9-HA-Ub conjugates signals are protected from external trypsin. Mitochondrial fractions were prepared from cells expressing HA-Ub or preSu9-HA-Ub, and mitochondrial fractions with and without trypsin treatment were subjected to western blot using α-HA antibodies. Tom70, a cytosol-facing mitochondrial outer membrane protein, was used as a marker exposed to the trypsin treatment, and Aco1 is a protected mitochondrial matrix marker. (E) preSu9-HA-Ub conjugates signals are not enhanced by proteasome inhibition treatment. Cytosolic (Cyto) and mitochondrial (Mito) subcellular fractions prepared from preSu9-HA-Ub expressing cells in the presence or absence of MG132 treatment were subjected to western blot. Detection of general and HA-tagged ubiquitin conjugates signals employed α-Ub and α-HA antibodies, respectively. Cytosolic marker Hxk1 and mitochondrial marker Aco1 are indicated.
) are indicated. (D) preSu9-HA-Ub conjugates signals are protected from external trypsin. Mitochondrial fractions were prepared from cells expressing HA-Ub or preSu9-HA-Ub, and mitochondrial fractions with and without trypsin treatment were subjected to western blot using α-HA antibodies. Tom70, a cytosol-facing mitochondrial outer membrane protein, was used as a marker exposed to the trypsin treatment, and Aco1 is a protected mitochondrial matrix marker. (E) preSu9-HA-Ub conjugates signals are not enhanced by proteasome inhibition treatment. Cytosolic (Cyto) and mitochondrial (Mito) subcellular fractions prepared from preSu9-HA-Ub expressing cells in the presence or absence of MG132 treatment were subjected to western blot. Detection of general and HA-tagged ubiquitin conjugates signals employed α-Ub and α-HA antibodies, respectively. Cytosolic marker Hxk1 and mitochondrial marker Aco1 are indicated.
 ), an outer membrane protein exposed to trypsin (
), an outer membrane protein exposed to trypsin ( ), a matrix protein protected inside mitochondria (
), a matrix protein protected inside mitochondria ( ), and trypsin (
), and trypsin ( ) are indicated. (D) preSu9-HA-Ub conjugates signals are protected from external trypsin. Mitochondrial fractions were prepared from cells expressing HA-Ub or preSu9-HA-Ub, and mitochondrial fractions with and without trypsin treatment were subjected to western blot using α-HA antibodies. Tom70, a cytosol-facing mitochondrial outer membrane protein, was used as a marker exposed to the trypsin treatment, and Aco1 is a protected mitochondrial matrix marker. (E) preSu9-HA-Ub conjugates signals are not enhanced by proteasome inhibition treatment. Cytosolic (Cyto) and mitochondrial (Mito) subcellular fractions prepared from preSu9-HA-Ub expressing cells in the presence or absence of MG132 treatment were subjected to western blot. Detection of general and HA-tagged ubiquitin conjugates signals employed α-Ub and α-HA antibodies, respectively. Cytosolic marker Hxk1 and mitochondrial marker Aco1 are indicated.
) are indicated. (D) preSu9-HA-Ub conjugates signals are protected from external trypsin. Mitochondrial fractions were prepared from cells expressing HA-Ub or preSu9-HA-Ub, and mitochondrial fractions with and without trypsin treatment were subjected to western blot using α-HA antibodies. Tom70, a cytosol-facing mitochondrial outer membrane protein, was used as a marker exposed to the trypsin treatment, and Aco1 is a protected mitochondrial matrix marker. (E) preSu9-HA-Ub conjugates signals are not enhanced by proteasome inhibition treatment. Cytosolic (Cyto) and mitochondrial (Mito) subcellular fractions prepared from preSu9-HA-Ub expressing cells in the presence or absence of MG132 treatment were subjected to western blot. Detection of general and HA-tagged ubiquitin conjugates signals employed α-Ub and α-HA antibodies, respectively. Cytosolic marker Hxk1 and mitochondrial marker Aco1 are indicated.







| IP | No. of Protein IDs a | No. of Mito Proteins | % of Mito Proteins b | No. of Matrix Proteins | % of Matrix Proteins c |
|---|---|---|---|---|---|
| HA-Ub | 557 | 269 | 48.3% | 69 | 25.7% |
| preSu9-HA-Ub | 131 | 109 | 83.2% | 78 | 71.6% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Karmon, O.; Das, K.; Wiener, R.; Lehming, N.; Pines, O. Ubiquitination Occurs in the Mitochondrial Matrix by Eclipsed Targeted Components of the Ubiquitination Machinery. Cells 2022, 11, 4109. https://doi.org/10.3390/cells11244109
Zhang Y, Karmon O, Das K, Wiener R, Lehming N, Pines O. Ubiquitination Occurs in the Mitochondrial Matrix by Eclipsed Targeted Components of the Ubiquitination Machinery. Cells. 2022; 11(24):4109. https://doi.org/10.3390/cells11244109
Chicago/Turabian StyleZhang, Yu, Ofri Karmon, Koyeli Das, Reuven Wiener, Norbert Lehming, and Ophry Pines. 2022. "Ubiquitination Occurs in the Mitochondrial Matrix by Eclipsed Targeted Components of the Ubiquitination Machinery" Cells 11, no. 24: 4109. https://doi.org/10.3390/cells11244109