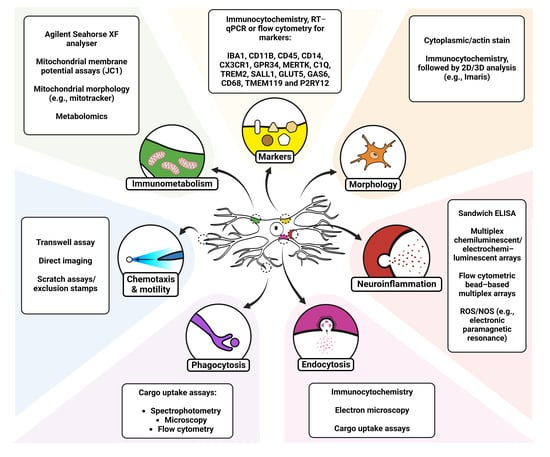
Assaying Microglia Functions In Vitro
Abstract
:1. Introduction
2. Markers for Microglia
3. Morphology
4. Neuroinflammation
4.1. Overview of Neuroinflammation
4.2. Stimuli Used in Neuroinflammation Assays
4.3. Functional Assays for Microglia-Mediated Neuroinflammation
5. Endocytosis
5.1. Overview of Endocytosis
5.2. Functional Assays for Endocytosis
6. Phagocytosis
6.1. Overview of Phagocytosis
6.2. Cargo Used in Microglia Phagocytosis Assays
6.3. General Considerations for Phagocytosis Assay Development
7. Chemotaxis and Random Mobility
7.1. Overview of Chemotaxis and Random Mobility
7.2. Functional Assays for Chemotaxis
Transwell Assays
7.3. Functional Assays for Random Motility
7.3.1. Direct Imaging
7.3.2. Scratch Assays/Exclusion Stamps
8. Microglial Immunometabolism
8.1. Overview of Microglial Immunometabolism
8.2. Functional Assays for Immunometabolism
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittelbronn, M.; Dietz, K.; Schluesener, H.J.; Meyermann, R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 2001, 101, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.L.; Yuan, Y.; Tian, L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 2020, 25, 351–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menassa, D.A.; Muntslag, T.A.O.; Martin-Estebané, M.; Barry-Carroll, L.; Chapman, M.A.; Adorjan, I.; Tyler, T.; Turnbull, B.; Rose-Zerilli, M.J.J.; Nicoll, J.A.R.; et al. The spatiotemporal dynamics of microglia across the human lifespan. Dev. Cell 2022, 57, 2127–2139.e6. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parakalan, R.; Jiang, B.; Nimmi, B.; Janani, M.; Jayapal, M.; Lu, J.; Tay, S.S.; Ling, E.A.; Dheen, S.T. Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 2012, 13, 64. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef]
- Ueno, M.; Fujita, Y.; Tanaka, T.; Nakamura, Y.; Kikuta, J.; Ishii, M.; Yamashita, T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013, 16, 543–551. [Google Scholar] [CrossRef]
- Hagemeyer, N.; Hanft, K.M.; Akriditou, M.A.; Unger, N.; Park, E.S.; Stanley, E.R.; Staszewski, O.; Dimou, L.; Prinz, M. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017, 134, 441–458. [Google Scholar] [CrossRef] [Green Version]
- Miron, V.E.; Boyd, A.; Zhao, J.W.; Yuen, T.J.; Ruckh, J.M.; Shadrach, J.L.; van Wijngaarden, P.; Wagers, A.J.; Williams, A.; Franklin, R.J.M.; et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 2013, 16, 1211–1218. [Google Scholar] [CrossRef] [Green Version]
- Marín-Teva, J.L.; Dusart, I.; Colin, C.; Gervais, A.; van Rooijen, N.; Mallat, M. Microglia promote the death of developing Purkinje cells. Neuron 2004, 41, 535–547. [Google Scholar] [CrossRef]
- Wakselman, S.; Béchade, C.; Roumier, A.; Bernard, D.; Triller, A.; Bessis, A. Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J. Neurosci. 2008, 28, 8138–8143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, C.L.; Martínez-Cerdeño, V.; Noctor, S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013, 33, 4216–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierra, A.; Encinas, J.M.; Deudero, J.J.; Chancey, J.H.; Enikolopov, G.; Overstreet-Wadiche, L.S.; Tsirka, S.E.; Maletic-Savatic, M. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 2010, 7, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Schafer, D.P.; Lehrman, E.K.; Kautzman, A.G.; Koyama, R.; Mardinly, A.R.; Yamasaki, R.; Ransohoff, R.M.; Greenberg, M.E.; Barres, B.A.; Stevens, B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012, 74, 691–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic pruning by microglia is necessary for normal brain development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshiko, M.; Arnoux, I.; Avignone, E.; Yamamoto, N.; Audinat, E. Deficiency of the microglial receptor CX3CR1 impairs postnatal functional development of thalamocortical synapses in the barrel cortex. J. Neurosci. 2012, 32, 15106–15111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pont-Lezica, L.; Beumer, W.; Colasse, S.; Drexhage, H.; Versnel, M.; Bessis, A. Microglia shape corpus callosum axon tract fasciculation: Functional impact of prenatal inflammation. Eur. J. Neurosci. 2014, 39, 1551–1557. [Google Scholar] [CrossRef]
- Squarzoni, P.; Oller, G.; Hoeffel, G.; Pont-Lezica, L.; Rostaing, P.; Low, D.; Bessis, A.; Ginhoux, F.; Garel, S. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014, 8, 1271–1279. [Google Scholar] [CrossRef] [Green Version]
- Augusto-Oliveira, M.; Arrifano, G.P.; Lopes-Araújo, A.; Santos-Sacramento, L.; Takeda, P.Y.; Anthony, D.C.; Malva, J.O.; Crespo-Lopez, M.E. What Do Microglia Really Do in Healthy Adult Brain? Cells 2019, 8, 1293. [Google Scholar] [CrossRef] [Green Version]
- Gumbs, S.B.H.; Kübler, R.; Gharu, L.; Schipper, P.J.; Borst, A.L.; Snijders, G.; Ormel, P.R.; van Berlekom, A.B.; Wensing, A.M.J.; de Witte, L.D.; et al. Human microglial models to study HIV infection and neuropathogenesis: A literature overview and comparative analyses. J. Neurovirol. 2022, 28, 64–91. [Google Scholar] [CrossRef]
- Timmerman, R.; Burm, S.M.; Bajramovic, J.J. An Overview of in vitro Methods to Study Microglia. Front. Cell Neurosci. 2018, 12, 242. [Google Scholar] [CrossRef] [PubMed]
- Cadiz, M.P.; Jensen, T.D.; Sens, J.P.; Zhu, K.; Song, W.M.; Zhang, B.; Ebbert, M.; Chang, R.; Fryer, J.D. Culture shock: Microglial heterogeneity, activation, and disrupted single-cell microglial networks in vitro. Mol. Neurodegener. 2022, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Dubbelaar, M.L.; Kracht, L.; Eggen, B.J.L.; Boddeke, E. The Kaleidoscope of Microglial Phenotypes. Front. Immunol. 2018, 9, 1753. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Prinz, M. Origin of microglia: Current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 2015, 7, a020537. [Google Scholar] [CrossRef] [Green Version]
- Böttcher, C.; Schlickeiser, S.; Sneeboer, M.A.M.; Kunkel, D.; Knop, A.; Paza, E.; Fidzinski, P.; Kraus, L.; Snijders, G.J.L.; Kahn, R.S.; et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 2019, 22, 78–90. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Weiner, H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef]
- Jurga, A.M.; Paleczna, M.; Kuter, K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell Neurosci. 2020, 14, 198. [Google Scholar] [CrossRef]
- Walker, D.G.; Tang, T.M.; Mendsaikhan, A.; Tooyama, I.; Serrano, G.E.; Sue, L.I.; Beach, T.G.; Lue, L.F. Patterns of Expression of Purinergic Receptor P2RY12, a Putative Marker for Non-Activated Microglia, in Aged and Alzheimer’s Disease Brains. Int. J. Mol. Sci. 2020, 21, 678. [Google Scholar] [CrossRef] [Green Version]
- Satoh, J.; Kino, Y.; Asahina, N.; Takitani, M.; Miyoshi, J.; Ishida, T.; Saito, Y. TMEM119 marks a subset of microglia in the human brain. Neuropathology 2016, 36, 39–49. [Google Scholar] [CrossRef]
- Hendrickx, D.A.E.; van Eden, C.G.; Schuurman, K.G.; Hamann, J.; Huitinga, I. Staining of HLA-DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. J. Neuroimmunol. 2017, 309, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.J.; Xie, H.; Zhang, C.Y.; Qin, H.F.; Zeng, X.W.; Lou, H.; Zhang, L.; Xu, G.T.; Zhang, J.F.; Xu, G.X. Is Iba-1 protein expression a sensitive marker for microglia activation in experimental diabetic retinopathy? Int. J. Ophthalmol. 2021, 14, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Honarpisheh, P.; Lee, J.; Banerjee, A.; Blasco-Conesa, M.P.; d’Aigle, J.; Mamun, A.A.; Ritzel, R.M.; Chauhan, A.; Ganesh, B.P.; McCullough, L.D. Potential caveats of putative microglia-specific markers for assessment of age-related cerebrovascular neuroinflammation. J. Neuroinflammation 2020, 17, 366. [Google Scholar] [CrossRef] [PubMed]
- Rangaraju, S.; Raza, S.A.; Li, N.X.; Betarbet, R.; Dammer, E.B.; Duong, D.; Lah, J.J.; Seyfried, N.T.; Levey, A.I. Differential Phagocytic Properties of CD45 (low) Microglia and CD45 (high) Brain Mononuclear Phagocytes-Activation and Age-Related Effects. Front. Immunol. 2018, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Bedard, K.; Sorce, S.; Hinz, B.; Dubois-Dauphin, M.; Krause, K.H. NOX4 expression in human microglia leads to constitutive generation of reactive oxygen species and to constitutive IL-6 expression. J. Innate. Immun. 2009, 1, 570–581. [Google Scholar] [CrossRef]
- Lier, J.; Streit, W.J.; Bechmann, I. Beyond Activation: Characterizing Microglial Functional Phenotypes. Cells 2021, 10, 2236. [Google Scholar] [CrossRef]
- Uff, C.E.G.; Patel, K.; Yeung, C.; Yip, P.K. Advances in Visualizing Microglial Cells in Human Central Nervous System Tissue. Biomolecules 2022, 12, 603. [Google Scholar] [CrossRef]
- Haynes, S.E.; Hollopeter, G.; Yang, G.; Kurpius, D.; Dailey, M.E.; Gan, W.B.; Julius, D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 2006, 9, 1512–1519. [Google Scholar] [CrossRef]
- Sousa, C.; Golebiewska, A.; Poovathingal, S.K.; Kaoma, T.; Pires-Afonso, Y.; Martina, S.; Coowar, D.; Azuaje, F.; Skupin, A.; Balling, R.; et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018, 19, e46171. [Google Scholar] [CrossRef]
- Hovens, I.; Nyakas, C.; Schoemaker, R. A novel method for evaluating microglial activation using ionized calcium-binding adaptor protein-1 staining: Cell body to cell size ratio. Neuroimmunol. Neuroinflammation 2014, 1, 82–88. [Google Scholar] [CrossRef]
- Dennis, J.; Morgan, M.K.; Graf, M.R.; Fuss, B. P2Y12 receptor expression is a critical determinant of functional responsiveness to ATX’s MORFO domain. Purinergic Signal. 2012, 8, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzeo, A.; Porta, M.; Beltramo, E. Characterization of an Immortalized Human Microglial Cell Line as a Tool for the Study of Diabetic Retinopathy. Int. J. Mol. Sci. 2022, 23, 5745. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.R.; Quan, Y.; Hanson, J.F.; Colonna, L.; Iorga, M.; Honda, S.; Shibuya, K.; Shibuya, A.; Elkon, K.B.; Möller, T. IgM-Dependent Phagocytosis in Microglia Is Mediated by Complement Receptor 3, Not Fcα/μ Receptor. J. Immunol. 2015, 195, 5309–5317. [Google Scholar] [CrossRef] [Green Version]
- Janda, E.; Boi, L.; Carta, A.R. Microglial Phagocytosis and Its Regulation: A Therapeutic Target in Parkinson’s Disease? Front. Mol. Neurosci. 2018, 11, 144. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Wang, K.; Hu, G.; Wang, X.; Miao, Z.; Azevedo, J.A.; Suh, E.; Van Deerlin, V.M.; Choi, D.; Roeder, K.; et al. APOE and TREM2 regulate amyloid-responsive microglia in Alzheimer’s disease. Acta Neuropathol. 2020, 140, 477–493. [Google Scholar] [CrossRef]
- Owens, R.; Grabert, K.; Davies, C.L.; Alfieri, A.; Antel, J.P.; Healy, L.M.; McColl, B.W. Divergent Neuroinflammatory Regulation of Microglial TREM Expression and Involvement of NF-κB. Front. Cell Neurosci. 2017, 11, 56. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, H.; Somoza, C.; Shigematsu, H.; Duprez, E.A.; Iwasaki-Arai, J.; Mizuno, S.; Arinobu, Y.; Geary, K.; Zhang, P.; Dayaram, T.; et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 2005, 106, 1590–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuehn, S.; Reinehr, S.; Stute, G.; Rodust, C.; Grotegut, P.; Hensel, A.T.; Dick, H.B.; Joachim, S.C. Interaction of complement system and microglia activation in retina and optic nerve in a NMDA damage model. Mol. Cell Neurosci. 2018, 89, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Brain Tissue Expression of MERTK—Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000153208-MERTK/brain (accessed on 5 October 2022).
- Chung, W.S.; Clarke, L.E.; Wang, G.X.; Stafford, B.K.; Sher, A.; Chakraborty, C.; Joung, J.; Foo, L.C.; Thompson, A.; Chen, C.; et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 2013, 504, 394–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healy, L.M.; Perron, G.; Won, S.Y.; Michell-Robinson, M.A.; Rezk, A.; Ludwin, S.K.; Moore, C.S.; Hall, J.A.; Bar-Or, A.; Antel, J.P. MerTK Is a Functional Regulator of Myelin Phagocytosis by Human Myeloid Cells. J. Immunol. 2016, 196, 3375–3384. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Buttgereit, A.; Lelios, I.; Yu, X.; Vrohlings, M.; Krakoski, N.R.; Gautier, E.L.; Nishinakamura, R.; Becher, B.; Greter, M. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 2016, 17, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pavlou, S.; Du, X.; Bhuckory, M.; Xu, H.; Chen, M. Glucose transporter 1 critically controls microglial activation through facilitating glycolysis. Mol. Neurodegener. 2019, 14, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilchrist, S.E.; Pennelli, G.M.; Hafizi, S. Gas6/TAM Signalling Negatively Regulates Inflammatory Induction of GM-CSF in Mouse Brain Microglia. Cells 2021, 10, 3281. [Google Scholar] [CrossRef]
- Liu, Y.; Walter, S.; Stagi, M.; Cherny, D.; Letiembre, M.; Schulz-Schaeffer, W.; Heine, H.; Penke, B.; Neumann, H.; Fassbender, K. LPS receptor (CD14): A receptor for phagocytosis of Alzheimer’s amyloid peptide. Brain 2005, 128, 1778–1789. [Google Scholar] [CrossRef] [Green Version]
- Sierra, A.; de Castro, F.; Del Río-Hortega, J.; Rafael Iglesias-Rozas, J.; Garrosa, M.; Kettenmann, H. The “Big-Bang” for modern glial biology: Translation and comments on Pío del Río-Hortega 1919 series of papers on microglia. Glia 2016, 64, 1801–1840. [Google Scholar] [CrossRef]
- Savage, J.C.; Carrier, M.; Tremblay, M. Morphology of Microglia Across Contexts of Health and Disease. Methods Mol. Biol. 2019, 2034, 13–26. [Google Scholar] [CrossRef]
- Stopper, L.; Bălşeanu, T.A.; Cătălin, B.; Rogoveanu, O.C.; Mogoantă, L.; Scheller, A. Microglia morphology in the physiological and diseased brain—From fixed tissue to in vivo conditions. Rom. J. Morphol. Embryol. 2018, 59, 7–12. [Google Scholar]
- Young, K.; Morrison, H. Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ. J. Vis. Exp. 2018, 136, 57648. [Google Scholar] [CrossRef] [Green Version]
- Bosch, L.F.P.; Kierdorf, K. The Shape of μ-How Morphological Analyses Shape the Study of Microglia. Front. Cell Neurosci. 2022, 16, 942462. [Google Scholar] [CrossRef]
- Althammer, F.; Ferreira-Neto, H.C.; Rubaharan, M.; Roy, R.K.; Patel, A.A.; Murphy, A.; Cox, D.N.; Stern, J.E. Three-dimensional morphometric analysis reveals time-dependent structural changes in microglia and astrocytes in the central amygdala and hypothalamic paraventricular nucleus of heart failure rats. J. Neuroinflammation 2020, 17, 221. [Google Scholar] [CrossRef]
- Srinivasan, A.; Ferland, R.J. AutoSholl allows for automation of Sholl analysis independent of user tracing. J. Neurosci. Methods 2020, 331, 108529. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.; Thompson-Peer, K.L. Comparing Automated Morphology Quantification Software on Dendrites of Uninjured and Injured Drosophila Neurons. Neuroinformatics 2021, 19, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Heindl, S.; Gesierich, B.; Benakis, C.; Llovera, G.; Duering, M.; Liesz, A. Automated Morphological Analysis of Microglia After Stroke. Front. Cell Neurosci. 2018, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Peng, H. APP2: Automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics 2013, 29, 1448–1454. [Google Scholar] [CrossRef]
- Megjhani, M.; Rey-Villamizar, N.; Merouane, A.; Lu, Y.; Mukherjee, A.; Trett, K.; Chong, P.; Harris, C.; Shain, W.; Roysam, B. Population-scale three-dimensional reconstruction and quantitative profiling of microglia arbors. Bioinformatics 2015, 31, 2190–2198. [Google Scholar] [CrossRef] [Green Version]
- York, E.M.; LeDue, J.M.; Bernier, L.P.; MacVicar, B.A. 3DMorph Automatic Analysis of Microglial Morphology in Three Dimensions from Ex Vivo and In Vivo Imaging. eNeuro 2018, 5, 0266-18. [Google Scholar] [CrossRef] [Green Version]
- Abdolhoseini, M.; Kluge, M.G.; Walker, F.R.; Johnson, S.J. Segmentation, Tracing, and Quantification of Microglial Cells from 3D Image Stacks. Sci. Rep. 2019, 9, 8557. [Google Scholar] [CrossRef] [Green Version]
- Davis, B.M.; Salinas-Navarro, M.; Cordeiro, M.F.; Moons, L.; De Groef, L. Characterizing microglia activation: A spatial statistics approach to maximize information extraction. Sci. Rep. 2017, 7, 1576. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Gómez, J.A.; Kavanagh, E.; Engskog-Vlachos, P.; Engskog, M.K.R.; Herrera, A.J.; Espinosa-Oliva, A.M.; Joseph, B.; Hajji, N.; Venero, J.L.; Burguillos, M.A. Microglia: Agents of the CNS Pro-Inflammatory Response. Cells 2020, 9, 1717. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Saresella, M.; La Rosa, F.; Piancone, F.; Zoppis, M.; Marventano, I.; Calabrese, E.; Rainone, V.; Nemni, R.; Mancuso, R.; Clerici, M. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol. Neurodegener 2016, 11, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [Green Version]
- Nizami, S.; Hall-Roberts, H.; Warrier, S.; Cowley, S.A.; Di Daniel, E. Microglial inflammation and phagocytosis in Alzheimer’s disease: Potential therapeutic targets. Br. J. Pharmacol. 2019, 176, 3515–3532. [Google Scholar] [CrossRef] [Green Version]
- Franco, R.; Fernández-Suárez, D. Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol. 2015, 131, 65–86. [Google Scholar] [CrossRef]
- Guo, S.; Wang, H.; Yin, Y. Microglia Polarization From M1 to M2 in Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 815347. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef]
- Lively, S.; Schlichter, L.C. Microglia Responses to Pro-inflammatory Stimuli (LPS, IFNγ+TNFα) and Reprogramming by Resolving Cytokines (IL-4, IL-10). Front. Cell Neurosci. 2018, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hou, B.; Liang, P.; Lu, X.; Wu, Y.; Zhang, X.; Fan, Y.; Liu, Y.; Chen, T.; Liu, W.; et al. TRPV1 channel mediates NLRP3 inflammasome-dependent neuroinflammation in microglia. Cell Death Dis. 2021, 12, 1159. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Leng, S.X.; McElhaney, J.E.; Walston, J.D.; Xie, D.; Fedarko, N.S.; Kuchel, G.A. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, B.; Dougherty, J.A.; Ponnalagu, D.; Singh, H.; Angelos, M.; Chen, C.A.; Khan, M. Measurement of Oxidative Stress Markers In Vitro Using Commercially Available Kits. In Measuring Oxidants and Oxidative Stress in Biological Systems; Berliner, L.J., Parinandi, N.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 39–60. [Google Scholar] [CrossRef]
- Domínguez-Punaro Mde, L.; Segura, M.; Contreras, I.; Lachance, C.; Houde, M.; Lecours, M.P.; Olivier, M.; Gottschalk, M. In vitro characterization of the microglial inflammatory response to Streptococcus suis, an important emerging zoonotic agent of meningitis. Infect Immun. 2010, 78, 5074–5085. [Google Scholar] [CrossRef] [Green Version]
- Nawab, A.; Nichols, A.; Klug, R.; Shapiro, J.I.; Sodhi, K. Spin Trapping: A Review for the Study of Obesity Related Oxidative Stress and Na(+)/K(+)-ATPase. J. Clin. Cell Immunol. 2017, 8, 505. [Google Scholar] [CrossRef] [Green Version]
- Solé-Domènech, S.; Cruz, D.L.; Capetillo-Zarate, E.; Maxfield, F.R. The endocytic pathway in microglia during health, aging and Alzheimer’s disease. Ageing Res. Rev. 2016, 32, 89–103. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Zhang, T.; Cheng, D.; Yang, J.; Chen, S.; Wang, X.; Li, X.; Duan, D.; Lou, H.; Zhu, L.; et al. Late endosomes promote microglia migration via cytosolic translocation of immature protease cathD. Sci. Adv. 2020, 6, eaba5783. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Anderson, R.G.; Brown, M.S. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 1979, 279, 679–685. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Lewis, W.H. Pinocytosis. Johns Hopkins Hosp. Bull. 1931, 49, 17–27. [Google Scholar]
- Chen, C.; Li, H.Q.; Liu, Y.J.; Guo, Z.F.; Wu, H.J.; Li, X.; Lou, H.F.; Zhu, L.; Wang, D.; Li, X.M.; et al. A novel size-based sorting mechanism of pinocytic luminal cargoes in microglia. J. Neurosci. 2015, 35, 2674–2688. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.C.; Teasdale, R.D. Defining macropinocytosis. Traffic 2009, 10, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Baranov, M.V.; Kumar, M.; Sacanna, S.; Thutupalli, S.; van den Bogaart, G. Modulation of Immune Responses by Particle Size and Shape. Front. Immunol. 2020, 11, 607945. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: The Legacy of Metchnikoff. Cell 2016, 166, 1065–1068. [Google Scholar] [CrossRef] [Green Version]
- Botté, A.; Lainé, J.; Xicota, L.; Heiligenstein, X.; Fontaine, G.; Kasri, A.; Rivals, I.; Goh, P.; Faklaris, O.; Cossec, J.C.; et al. Ultrastructural and dynamic studies of the endosomal compartment in Down syndrome. Acta Neuropathol. Commun. 2020, 8, 89. [Google Scholar] [CrossRef]
- Bonet-Ponce, L.; Beilina, A.; Williamson, C.D.; Lindberg, E.; Kluss, J.H.; Saez-Atienzar, S.; Landeck, N.; Kumaran, R.; Mamais, A.; Bleck, C.K.E.; et al. LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci. Adv. 2020, 6, eabb2454. [Google Scholar] [CrossRef]
- Knupp, A.; Mishra, S.; Martinez, R.; Braggin, J.E.; Szabo, M.; Kinoshita, C.; Hailey, D.W.; Small, S.A.; Jayadev, S.; Young, J.E. Depletion of the AD Risk Gene SORL1 Selectively Impairs Neuronal Endosomal Traffic Independent of Amyloidogenic APP Processing. Cell Rep. 2020, 31, 107719. [Google Scholar] [CrossRef]
- Kwart, D.; Gregg, A.; Scheckel, C.; Murphy, E.A.; Paquet, D.; Duffield, M.; Fak, J.; Olsen, O.; Darnell, R.B.; Tessier-Lavigne, M. A Large Panel of Isogenic APP and PSEN1 Mutant Human iPSC Neurons Reveals Shared Endosomal Abnormalities Mediated by APP β-CTFs, Not Aβ. Neuron 2019, 104, 256–270.e5. [Google Scholar] [CrossRef]
- Cataldo, A.M.; Peterhoff, C.M.; Troncoso, J.C.; Gomez-Isla, T.; Hyman, B.T.; Nixon, R.A. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: Differential effects of APOE genotype and presenilin mutations. Am. J. Pathol. 2000, 157, 277–286. [Google Scholar] [CrossRef]
- Small, S.A.; Simoes-Spassov, S.; Mayeux, R.; Petsko, G.A. Endosomal Traffic Jams Represent a Pathogenic Hub and Therapeutic Target in Alzheimer’s Disease. Trends Neurosci. 2017, 40, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, Z.P.; Bretou, M.; Annaert, W. Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: Impact of genetic risk factors. Mol. Neurodegener. 2019, 14, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, M.P.; Mishra, S.; Knupp, A.; Young, J.E. The role of Alzheimer’s disease risk genes in endolysosomal pathways. Neurobiol. Dis. 2022, 162, 105576. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Redpath, G.M.I.; Nicovich, P.R.; Rossy, J. Quantitative visualization of endocytic trafficking through photoactivation of fluorescent proteins. Mol. Biol. Cell 2021, 32, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Dräger, N.M.; Sattler, S.M.; Huang, C.T.; Teter, O.M.; Leng, K.; Hashemi, S.H.; Hong, J.; Aviles, G.; Clelland, C.D.; Zhan, L.; et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022, 25, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.E.; Colombo, G.; Schulz, R.; Siegert, S. Targeting microglia with lentivirus and AAV: Recent advances and remaining challenges. Neurosci. Lett. 2019, 707, 134310. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, S.; Jiang, Q.; Lee, C.Y.; Koenigsknecht-Talboo, J.; Holtzman, D.M.; Landreth, G.E. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J. Neurosci. 2009, 29, 4252–4262. [Google Scholar] [CrossRef] [Green Version]
- Mamais, A.; Kluss, J.H.; Bonet-Ponce, L.; Landeck, N.; Langston, R.G.; Smith, N.; Beilina, A.; Kaganovich, A.; Ghosh, M.C.; Pellegrini, L.; et al. Mutations in LRRK2 linked to Parkinson disease sequester Rab8a to damaged lysosomes and regulate transferrin-mediated iron uptake in microglia. PLoS Biol. 2021, 19, e3001480. [Google Scholar] [CrossRef]
- Narayan, P.; Sienski, G.; Bonner, J.M.; Lin, Y.T.; Seo, J.; Baru, V.; Haque, A.; Milo, B.; Akay, L.A.; Graziosi, A.; et al. PICALM Rescues Endocytic Defects Caused by the Alzheimer’s Disease Risk Factor APOE4. Cell Rep. 2020, 33, 108224. [Google Scholar] [CrossRef]
- Connor-Robson, N.; Booth, H.; Martin, J.G.; Gao, B.; Li, K.; Doig, N.; Vowles, J.; Browne, C.; Klinger, L.; Juhasz, P.; et al. An integrated transcriptomics and proteomics analysis reveals functional endocytic dysregulation caused by mutations in LRRK2. Neurobiol. Dis. 2019, 127, 512–526. [Google Scholar] [CrossRef]
- Wang, J.T.; Teasdale, R.D.; Liebl, D. Macropinosome quantitation assay. MethodsX 2014, 1, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Kleist, L.; Stahlschmidt, W.; Bulut, H.; Gromova, K.; Puchkov, D.; Robertson, M.J.; MacGregor, K.A.; Tomilin, N.; Pechstein, A.; Chau, N.; et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 2011, 146, 471–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, S.L.; Korn, E.D. Substoichiometric concentrations of cytochalasin D inhibit actin polymerization. Additional evidence for an F-actin treadmill. J. Biol. Chem. 1979, 254, 9982–9985. [Google Scholar] [CrossRef]
- Gladhaug, I.P.; Christoffersen, T. Amiloride inhibits constitutive internalization and increases the surface number of epidermal growth factor receptors in intact rat hepatocytes. J. Cell Physiol. 1990, 143, 188–195. [Google Scholar] [CrossRef]
- Scheiblich, H.; Dansokho, C.; Mercan, D.; Schmidt, S.V.; Bousset, L.; Wischhof, L.; Eikens, F.; Odainic, A.; Spitzer, J.; Griep, A.; et al. Microglia jointly degrade fibrillar alpha-synuclein cargo by distribution through tunneling nanotubes. Cell 2021, 184, 5089–5106.e21. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, K.S.; Lee, S.B.; Ryu, J.S.; Chung, K.C.; Choo, Y.K.; Jou, I.; Kim, J.; Park, S.M. On the mechanism of internalization of alpha-synuclein into microglia: Roles of ganglioside GM1 and lipid raft. J. Neurochem. 2009, 110, 400–411. [Google Scholar] [CrossRef]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 2018, 98, 1141–1154.e7. [Google Scholar] [CrossRef] [Green Version]
- Maguire, E.; Menzies, G.E.; Phillips, T.; Sasner, M.; Williams, H.M.; Czubala, M.A.; Evans, N.; Cope, E.L.; Sims, R.; Howell, G.R.; et al. PIP2 depletion and altered endocytosis caused by expression of Alzheimer’s disease-protective variant PLCγ2 R522. Embo J. 2021, 40, e105603. [Google Scholar] [CrossRef]
- Wells, C.; Brennan, S.; Keon, M.; Ooi, L. The role of amyloid oligomers in neurodegenerative pathologies. Int. J. Biol. Macromol. 2021, 181, 582–604. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e9. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Ettle, B.; Bruno, A.; Kulinich, A.; Hoffmann, A.C.; von Wittgenstein, J.; Winkler, J.; Xiang, W.; Schlachetzki, J.C.M. Alpha-synuclein activates BV2 microglia dependent on its aggregation state. Biochem. Biophys. Res. Commun. 2016, 479, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Sondag, C.M.; Dhawan, G.; Combs, C.K. Beta amyloid oligomers and fibrils stimulate differential activation of primary microglia. J. Neuroinflammation 2009, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ries, M.; Sastre, M. Mechanisms of Aβ Clearance and Degradation by Glial Cells. Front. Aging Neurosci. 2016, 8, 160. [Google Scholar] [CrossRef] [Green Version]
- Joshi, P.; Riffel, F.; Satoh, K.; Enomoto, M.; Qamar, S.; Scheiblich, H.; Villacampa, N.; Kumar, S.; Theil, S.; Parhizkar, S.; et al. Differential interaction with TREM2 modulates microglial uptake of modified Aβ species. Glia 2021, 69, 2917–2932. [Google Scholar] [CrossRef]
- Das, R.; Balmik, A.A.; Chinnathambi, S. Phagocytosis of full-length Tau oligomers by Actin-remodeling of activated microglia. J. Neuroinflammation 2020, 17, 10. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.A.; Grinstein, S. Phagocytosis: Receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014, 262, 193–215. [Google Scholar] [CrossRef]
- Kono, R.; Ikegaya, Y.; Koyama, R. Phagocytic Glial Cells in Brain Homeostasis. Cells 2021, 10, 1348. [Google Scholar] [CrossRef]
- Fountain, A.; Inpanathan, S.; Alves, P.; Verdawala, M.B.; Botelho, R.J. Phagosome maturation in macrophages: Eat, digest, adapt, and repeat. Adv. Biol. Regul. 2021, 82, 100832. [Google Scholar] [CrossRef]
- Bhattacherjee, A.; Daskhan, G.C.; Bains, A.; Watson, A.E.S.; Eskandari-Sedighi, G.; St Laurent, C.D.; Voronova, A.; Macauley, M.S. Increasing phagocytosis of microglia by targeting CD33 with liposomes displaying glycan ligands. J. Control. Release 2021, 338, 680–693. [Google Scholar] [CrossRef]
- Garcia-Reitboeck, P.; Phillips, A.; Piers, T.M.; Villegas-Llerena, C.; Butler, M.; Mallach, A.; Rodrigues, C.; Arber, C.E.; Heslegrave, A.; Zetterberg, H.; et al. Human Induced Pluripotent Stem Cell-Derived Microglia-Like Cells Harboring TREM2 Missense Mutations Show Specific Deficits in Phagocytosis. Cell Rep. 2018, 24, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Flynn, R.; Sharma, I.; Haberman, E.; Carling, P.J.; Nicholls, F.J.; Stegmann, M.; Vowles, J.; Haenseler, W.; Wade-Martins, R.; et al. LRRK2 Is Recruited to Phagosomes and Co-recruits RAB8 and RAB10 in Human Pluripotent Stem Cell-Derived Macrophages. Stem Cell Rep. 2020, 14, 940–955. [Google Scholar] [CrossRef]
- Butler, C.A.; Popescu, A.S.; Kitchener, E.J.A.; Allendorf, D.H.; Puigdellívol, M.; Brown, G.C. Microglial phagocytosis of neurons in neurodegeneration, and its regulation. J. Neurochem. 2021, 158, 621–639. [Google Scholar] [CrossRef]
- Neniskyte, U.; Neher, J.J.; Brown, G.C. Neuronal death induced by nanomolar amyloid β is mediated by primary phagocytosis of neurons by microglia. J. Biol. Chem. 2011, 286, 39904–39913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muth, C.; Hartmann, A.; Sepulveda-Falla, D.; Glatzel, M.; Krasemann, S. Phagocytosis of Apoptotic Cells Is Specifically Upregulated in ApoE4 Expressing Microglia in vitro. Front. Cell Neurosci. 2019, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Hall-Roberts, H.; Agarwal, D.; Obst, J.; Smith, T.B.; Monzón-Sandoval, J.; Di Daniel, E.; Webber, C.; James, W.S.; Mead, E.; Davis, J.B.; et al. TREM2 Alzheimer’s variant R47H causes similar transcriptional dysregulation to knockout, yet only subtle functional phenotypes in human iPSC-derived macrophages. Alzheimers Res. Ther. 2020, 12, 151. [Google Scholar] [CrossRef]
- Atagi, Y.; Liu, C.C.; Painter, M.M.; Chen, X.F.; Verbeeck, C.; Zheng, H.; Li, X.; Rademakers, R.; Kang, S.S.; Xu, H.; et al. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 2015, 290, 26043–26050. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Rochford, C.D.; Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005, 201, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Kawabori, M.; Kacimi, R.; Kauppinen, T.; Calosing, C.; Kim, J.Y.; Hsieh, C.L.; Nakamura, M.C.; Yenari, M.A. Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J. Neurosci. 2015, 35, 3384–3396. [Google Scholar] [CrossRef] [Green Version]
- Rios-Doria, J.; Favata, M.; Lasky, K.; Feldman, P.; Lo, Y.; Yang, G.; Stevens, C.; Wen, X.; Sehra, S.; Katiyar, K.; et al. A Potent and Selective Dual Inhibitor of AXL and MERTK Possesses Both Immunomodulatory and Tumor-Targeted Activity. Front. Oncol. 2020, 10, 598477. [Google Scholar] [CrossRef]
- Reichert, F.; Rotshenker, S. Complement-receptor-3 and scavenger-receptor-AI/II mediated myelin phagocytosis in microglia and macrophages. Neurobiol. Dis. 2003, 12, 65–72. [Google Scholar] [CrossRef]
- Andreone, B.J.; Przybyla, L.; Llapashtica, C.; Rana, A.; Davis, S.S.; van Lengerich, B.; Lin, K.; Shi, J.; Mei, Y.; Astarita, G.; et al. Alzheimer’s-associated PLCγ2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nat. Neurosci. 2020, 23, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, D.A.; Schuurman, K.G.; van Draanen, M.; Hamann, J.; Huitinga, I. Enhanced uptake of multiple sclerosis-derived myelin by THP-1 macrophages and primary human microglia. J. Neuroinflammation 2014, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Sokolova, D.; Childs, T.; Hong, S. Insight into the role of phosphatidylserine in complement-mediated synapse loss in Alzheimer’s disease. Fac. Rev. 2021, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Weinhard, L.; di Bartolomei, G.; Bolasco, G.; Machado, P.; Schieber, N.L.; Neniskyte, U.; Exiga, M.; Vadisiute, A.; Raggioli, A.; Schertel, A.; et al. Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun. 2018, 9, 1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gylys, K.H.; Fein, J.A.; Yang, F.; Cole, G.M. Enrichment of presynaptic and postsynaptic markers by size-based gating analysis of synaptosome preparations from rat and human cortex. Cytom. A 2004, 60, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bido, S.; Muggeo, S.; Massimino, L.; Marzi, M.J.; Giannelli, S.G.; Melacini, E.; Nannoni, M.; Gambarè, D.; Bellini, E.; Ordazzo, G.; et al. Microglia-specific overexpression of α-synuclein leads to severe dopaminergic neurodegeneration by phagocytic exhaustion and oxidative toxicity. Nat. Commun. 2021, 12, 6237. [Google Scholar] [CrossRef]
- Sellgren, C.M.; Sheridan, S.D.; Gracias, J.; Xuan, D.; Fu, T.; Perlis, R.H. Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol. Psychiatry 2017, 22, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Benetatos, J.; Bennett, R.E.; Evans, H.T.; Ellis, S.A.; Hyman, B.T.; Bodea, L.G.; Götz, J. PTEN activation contributes to neuronal and synaptic engulfment by microglia in tauopathy. Acta Neuropathol. 2020, 140, 7–24. [Google Scholar] [CrossRef] [Green Version]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef]
- Bohlen, C.J.; Bennett, F.C.; Tucker, A.F.; Collins, H.Y.; Mulinyawe, S.B.; Barres, B.A. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 2017, 94, 759–773.e8. [Google Scholar] [CrossRef] [PubMed]
- Hall-Roberts, H.; Di Daniel, E.; James, W.S.; Davis, J.B.; Cowley, S.A. In Vitro Quantitative Imaging Assay for Phagocytosis of Dead Neuroblastoma Cells by iPSC-Macrophages. J. Vis. Exp. 2021, 168, e62217. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.J.; Sun, C.; Fuller, S.; Skarratt, K.K.; Petrou, S.; Wiley, J.S. A quantitative method for measuring innate phagocytosis by human monocytes using real-time flow cytometry. Cytom. A 2014, 85, 313–321. [Google Scholar] [CrossRef]
- Caldeira, C.; Oliveira, A.F.; Cunha, C.; Vaz, A.R.; Falcão, A.S.; Fernandes, A.; Brites, D. Microglia change from a reactive to an age-like phenotype with the time in culture. Front. Cell Neurosci. 2014, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Miksa, M.; Komura, H.; Wu, R.; Shah, K.G.; Wang, P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 2009, 342, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes-Hasler, P.; Maschalidi, S.; Lippens, C.; Castelbou, C.; Bouvet, S.; Guido, D.; Bermont, F.; Bassoy, E.Y.; Page, N.; Merkler, D.; et al. STIM1 promotes migration, phagosomal maturation and antigen cross-presentation in dendritic cells. Nat. Commun. 2017, 8, 1852. [Google Scholar] [CrossRef] [Green Version]
- Nuutila, J.; Lilius, E.M. Flow cytometric quantitative determination of ingestion by phagocytes needs the distinguishing of overlapping populations of binding and ingesting cells. Cytom. A 2005, 65, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Madry, C.; Kyrargyri, V.; Arancibia-Cárcamo, I.L.; Jolivet, R.; Kohsaka, S.; Bryan, R.M.; Attwell, D. Microglial Ramification, Surveillance, and Interleukin-1β Release Are Regulated by the Two-Pore Domain K(+) Channel THIK-1. Neuron 2018, 97, 299–312.e6. [Google Scholar] [CrossRef]
- Frow, E.K.; Reckless, J.; Grainger, D.J. Tools for anti-inflammatory drug design: In vitro models of leukocyte migration. Med. Res. Rev. 2004, 24, 276–298. [Google Scholar] [CrossRef]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef]
- Miller, E.B.; Zhang, P.; Ching, K.; Pugh, E.N., Jr.; Burns, M.E. In vivo imaging reveals transient microglia recruitment and functional recovery of photoreceptor signaling after injury. Proc. Natl. Acad. Sci. USA 2019, 116, 16603–16612. [Google Scholar] [CrossRef] [PubMed]
- Isfort, K.; Ebert, F.; Bornhorst, J.; Sargin, S.; Kardakaris, R.; Pasparakis, M.; Bähler, M.; Schwerdtle, T.; Schwab, A.; Hanley, P.J. Real-time imaging reveals that P2Y2 and P2Y12 receptor agonists are not chemoattractants and macrophage chemotaxis to complement C5a is phosphatidylinositol 3-kinase (PI3K)- and p38 mitogen-activated protein kinase (MAPK)-independent. J. Biol. Chem. 2011, 286, 44776–44787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumianek, A.N.; Greaves, D.R. How Have Leukocyte In Vitro Chemotaxis Assays Shaped Our Ideas about Macrophage Migration? Biology 2020, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Sasaki, Y.; Ohsawa, K.; Imai, Y.; Nakamura, Y.; Inoue, K.; Kohsaka, S. Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J. Neurosci. 2001, 21, 1975–1982. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Hashimoto, T.; Wong, E.; Hori, Y.; Wood, L.B.; Zhao, L.; Haigis, K.M.; Hyman, B.T.; Irimia, D. Microfluidic chemotaxis platform for differentiating the roles of soluble and bound amyloid-β on microglial accumulation. Sci. Rep. 2013, 3, 1823. [Google Scholar] [CrossRef] [Green Version]
- Lelli, A.; Gervais, A.; Colin, C.; Chéret, C.; Ruiz de Almodovar, C.; Carmeliet, P.; Krause, K.H.; Boillée, S.; Mallat, M. The NADPH oxidase Nox2 regulates VEGFR1/CSF-1R-mediated microglial chemotaxis and promotes early postnatal infiltration of phagocytes in the subventricular zone of the mouse cerebral cortex. Glia 2013, 61, 1542–1555. [Google Scholar] [CrossRef]
- Zhang, J.; Gong, X.; Xiong, H. Significant higher-level C-C motif chemokine ligand 2/3 and chemotactic power in cerebral white matter than grey matter in rat and human. Eur. J. Neurosci. 2021, 54, 4088–4100. [Google Scholar] [CrossRef]
- Das, R.; Chinnathambi, S. Microglial remodeling of actin network by Tau oligomers, via G protein-coupled purinergic receptor, P2Y12R-driven chemotaxis. Traffic 2021, 22, 153–170. [Google Scholar] [CrossRef]
- Wang, S.; Chu, C.H.; Stewart, T.; Ginghina, C.; Wang, Y.; Nie, H.; Guo, M.; Wilson, B.; Hong, J.S.; Zhang, J. α-Synuclein, a chemoattractant, directs microglial migration via H2O2-dependent Lyn phosphorylation. Proc. Natl. Acad. Sci. USA 2015, 112, E1926–E1935. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.K.; Cho, T.; Choi, H.B.; Wang, Y.T.; McLarnon, J.G. Microglial VEGF receptor response is an integral chemotactic component in Alzheimer’s disease pathology. J. Neurosci. 2009, 29, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Haenseler, W.; Sansom, S.N.; Buchrieser, J.; Newey, S.E.; Moore, C.S.; Nicholls, F.J.; Chintawar, S.; Schnell, C.; Antel, J.P.; Allen, N.D.; et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Rep. 2017, 8, 1727–1742. [Google Scholar] [CrossRef]
- Kyrargyri, V.; Attwell, D.; Jolivet, R.B.; Madry, C. Analysis of Signaling Mechanisms Regulating Microglial Process Movement. Methods Mol. Biol. 2019, 2034, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Hefendehl, J.K.; Neher, J.J.; Sühs, R.B.; Kohsaka, S.; Skodras, A.; Jucker, M. Homeostatic and injury-induced microglia behavior in the aging brain. Aging Cell 2014, 13, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Jairaman, A.; McQuade, A.; Granzotto, A.; Kang, Y.J.; Chadarevian, J.P.; Gandhi, S.; Parker, I.; Smith, I.; Cho, H.; Sensi, S.L.; et al. TREM2 regulates purinergic receptor-mediated calcium signaling and motility in human iPSC-derived microglia. Elife 2022, 11, e73021. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robichon, K.; Patel, V.; Connor, B.; La Flamme, A.C. Clozapine reduces infiltration into the CNS by targeting migration in experimental autoimmune encephalomyelitis. J. Neuroinflammation 2020, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Fairley, L.H.; Wong, J.H.; Barron, A.M. Mitochondrial Regulation of Microglial Immunometabolism in Alzheimer’s Disease. Front. Immunol. 2021, 12, 624538. [Google Scholar] [CrossRef]
- Culmsee, C.; Michels, S.; Scheu, S.; Arolt, V.; Dannlowski, U.; Alferink, J. Mitochondria, Microglia, and the Immune System-How Are They Linked in Affective Disorders? Front. Psychiatry 2018, 9, 739. [Google Scholar] [CrossRef] [Green Version]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef]
- Pereira, O.R., Jr.; Ramos, V.M.; Cabral-Costa, J.V.; Kowaltowski, A.J. Changes in mitochondrial morphology modulate LPS-induced loss of calcium homeostasis in BV-2 microglial cells. J. Bioenerg. Biomembr. 2021, 53, 109–118. [Google Scholar] [CrossRef]
- Park, J.; Choi, H.; Kim, B.; Chae, U.; Lee, D.G.; Lee, S.R.; Lee, S.; Lee, H.S.; Lee, D.S. Peroxiredoxin 5 (Prx5) decreases LPS-induced microglial activation through regulation of Ca(2+)/calcineurin-Drp1-dependent mitochondrial fission. Free Radic. Biol. Med. 2016, 99, 392–404. [Google Scholar] [CrossRef]
- Park, J.; Choi, H.; Min, J.S.; Park, S.J.; Kim, J.H.; Park, H.J.; Kim, B.; Chae, J.I.; Yim, M.; Lee, D.S. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 2013, 127, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Castillo, E.; Frias, E.S.; Swanson, R.A. Bioenergetic regulation of microglia. Glia 2018, 66, 1200–1212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piers, T.M.; Cosker, K.; Mallach, A.; Johnson, G.T.; Guerreiro, R.; Hardy, J.; Pocock, J.M. A locked immunometabolic switch underlies TREM2 R47H loss of function in human iPSC-derived microglia. FASEB J. 2020, 34, 2436–2450. [Google Scholar] [CrossRef] [Green Version]
- Kalsbeek, M.J.; Mulder, L.; Yi, C.X. Microglia energy metabolism in metabolic disorder. Mol. Cell Endocrinol. 2016, 438, 27–35. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaguchi, H.; Horikoshi, Y.; Tanaka, G.; Nakazato, Y. Expression of glucose transporter 5 by microglia in human gliomas. Neuropathol. Appl. Neurobiol. 2004, 30, 447–455. [Google Scholar] [CrossRef]
- Brusco, J.; Haas, K. Interactions between mitochondria and the transcription factor myocyte enhancer factor 2 (MEF2) regulate neuronal structural and functional plasticity and metaplasticity. J. Physiol. 2015, 593, 3471–3481. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.; Sun, X.; Wang, P.; Zhang, S.; Wang, X.; Wu, H.; Hong, L.; Xie, C.; Li, X.; Zhao, H.; et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat. Immunol. 2015, 16, 1142–1152. [Google Scholar] [CrossRef] [Green Version]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.; Sobotka, K.S.; Joshi, P.; Gressens, P.; Fleiss, B.; Thornton, C.; Mallard, C.; Hagberg, H. Lipopolysaccharide-induced alteration of mitochondrial morphology induces a metabolic shift in microglia modulating the inflammatory response in vitro and in vivo. Glia 2019, 67, 1047–1061. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.M.; Liu, N.; Qin, Z.H.; Wang, Y. Mitochondrial-derived damage-associated molecular patterns amplify neuroinflammation in neurodegenerative diseases. Acta Pharmacol. Sin. 2022, 43, 2439–2447. [Google Scholar] [CrossRef]
- Voss, K.; Hong, H.S.; Bader, J.E.; Sugiura, A.; Lyssiotis, C.A.; Rathmell, J.C. A guide to interrogating immunometabolism. Nat. Rev. Immunol. 2021, 21, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Mookerjee, S.A.; Gerencser, A.A.; Nicholls, D.G.; Brand, M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 2017, 292, 7189–7207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, N.M.C.; Theurey, P.; Adam-Vizi, V.; Bazan, N.G.; Bernardi, P.; Bolaños, J.P.; Culmsee, C.; Dawson, V.L.; Deshmukh, M.; Duchen, M.R.; et al. Guidelines on experimental methods to assess mitochondrial dysfunction in cellular models of neurodegenerative diseases. Cell Death Differ. 2018, 25, 542–572. [Google Scholar] [CrossRef] [Green Version]
- Scaduto, R.C., Jr.; Grotyohann, L.W. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys. J. 1999, 76, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Glancy, B.; Kim, Y.; Katti, P.; Willingham, T.B. The Functional Impact of Mitochondrial Structure Across Subcellular Scales. Front. Physiol. 2020, 11, 541040. [Google Scholar] [CrossRef]



| Marker | Function | Expression in CNS Cell Types and Microglial Models | Expression after Proinflammatory Stimulation | References |
|---|---|---|---|---|
| TMEM119 | Currently uncertain | Microglia. Highly conserved. Low mRNA in some iPS models. | Downregulated | [26,30,39] |
| IBA1 | Binds calcium and actin supporting the cytoskeleton and membrane ruffling | Microglia. | Upregulated | [40] |
| P2RY12 | Detects nucleotides such a ATP following injury | Microglia and oligodendrocyte precursor cells. Low mRNA in some iPS models. | Downregulated | [26,38,41] |
| CD11B | An integrin for complement receptor 3 (CR3), used for phagocytosis of complement-coated cargo | Microglia. Some have shown to be absent on the HMC3 cell line. | Upregulated | [34,42,43] |
| CD45 | Receptor protein tyrosine phosphate involved with T cell signalling and proliferation | Microglia. | Upregulated | [34,35] |
| CD68 | Lysosomal marker | Microglia. Some have shown to be absent on the HMC3 cell line. | Unchanged | [26,42,44] |
| TREM2 | Key in development and maintenance of the brain with a role in synaptic pruning and immune response | Microglia. | Downregulated | [34,39,45,46] |
| PU.1 | Transcription factor determining myeloid lineage | Microglia. | Unchanged | [47] |
| C1QA | Opsonin that triggers the classical complement cascade | Microglia. | Upregulated | [26,48] |
| MERTK | Transduces signals, involved in cell survival, migration and phagocytosis | Microglia, astrocytes, rod photoreceptor cells and oligodendrocyte precursor cell. | Downregulated | [39,49,50,51] |
| CX3CR1 | Involved in the immune response, inflammation, cell adhesion, chemotaxis and migration | Microglia. | Upregulated | [26,52] |
| SALL1 | Transcriptional master regulator of microglia identity and non-inflammatory functions | Microglia, oligodendrocytes, and astrocytes. | Downregulated | [27,39,53] |
| GLUT5 | Fructose transporter | Microglia and some neuronal populations. | Upregulated | [54] |
| GAS6 | Simulates cell proliferation and shown to blunt inflammatory response of LPS | Microglia, fibroblasts and astrocytes. | No information available | [55] |
| CD14 | Co receptor for TLR4 and TLR7/9 | Microglia. | Upregulated | [26,56] |
| Type of Assay | Max Cytokines/Assay | Pros | Cons | Examples |
|---|---|---|---|---|
| Double antibody sandwich ELISA | 1 | Most widely used & best validated, highly quantitative, reproducible | Measures only a single protein/sample, dynamic range narrow in relation to other cytokine assays | R&D Systems® ELISA Kits, Abcam ELISA Kits, Invitrogen ELISA kits |
| Multiplex chemiluminescent arrays | 9 | Efficiency (time & cost), easier to do multiple timepoints, higher dynamic range than traditional ELISA, less sample needed, allows evaluation of one inflammatory molecule in the context of multiple others | May require specialist paid software to analyse results (e.g., Quansys Q-plex™ requires Q-view software) | Quansys Q-Plex™ (Oxford Biosystems), Luminex® Multiplex Assays (Thermofisher), Multiplex Immunoassays (Bio-Rad) |
| Multiplex electrochemiluminescence arrays | 10 | Efficiency (time & cost), lack of enzymatic or fluorescent detection system avoids time-dependent signal decay, less sample needed, allows evaluation of one inflammatory molecule in the context of multiple others | Expensive, requires specialist equipment | Meso Scale Discovery (MSD) |
| Flow cytometric bead-based multiplex arrays | 100 | Efficiency (time & cost), compatible with standard flow cytometers, allows evaluation of one inflammatory molecule in the context of multiple others, free analysis software, cheapest method per sample/cytokine, easier to do multiple timepoints, less sample needed | Less tested compared to traditional sandwich ELISA | BD Cytometric Bead Array (BD Biosciences), LEGENDplex™ (BioLegend) |
| Spectro-Photometer | Fluorescence Microscope (No Automation) | Time-Lapse Fluorescence Microscope | High-Content Imaging System | Flow Cytometer | Imaging Flow Cytometer | |
|---|---|---|---|---|---|---|
| Sensitivity | + | ID | ++ | +++ | +++ | +++ |
| Magnification | N/A | ID | ++ | +++ | N/A | ++ |
| Multiple data parameters | + | ID | ++ | +++ | + | ++ |
| Unbiased data collection | +++ | + | ++ | +++ | +++ | +++ |
| Unbiased data analysis | +++ | + | ++ | +++ | ++ | ++ |
| Speed | +++ | + | ++ | +++ | + | + |
| Cell viability | ++ | ++ | +++ | +++ | + | + |
| Real-time kinetics | Y | N/A | Y | Y | N/A | N/A |
| Single-cell analysis | N/A | Y | ID | Y | Y | Y |
| Example instrument | SpectraMax (Molecular Devices) | LSM700 (Zeiss) | Incucyte ZOOM (Sartorius) | Opera Phenix (Perkin Elmer) | FACS Calibur (Becton Dickinson) | Amnis ImageStreamX Mk II (Luminex) |
| Assay references | [151] | [152] | [153] | [138,144] | [133,145] | [123,137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maguire, E.; Connor-Robson, N.; Shaw, B.; O’Donoghue, R.; Stöberl, N.; Hall-Roberts, H. Assaying Microglia Functions In Vitro. Cells 2022, 11, 3414. https://doi.org/10.3390/cells11213414
Maguire E, Connor-Robson N, Shaw B, O’Donoghue R, Stöberl N, Hall-Roberts H. Assaying Microglia Functions In Vitro. Cells. 2022; 11(21):3414. https://doi.org/10.3390/cells11213414
Chicago/Turabian StyleMaguire, Emily, Natalie Connor-Robson, Bethany Shaw, Rachel O’Donoghue, Nina Stöberl, and Hazel Hall-Roberts. 2022. "Assaying Microglia Functions In Vitro" Cells 11, no. 21: 3414. https://doi.org/10.3390/cells11213414